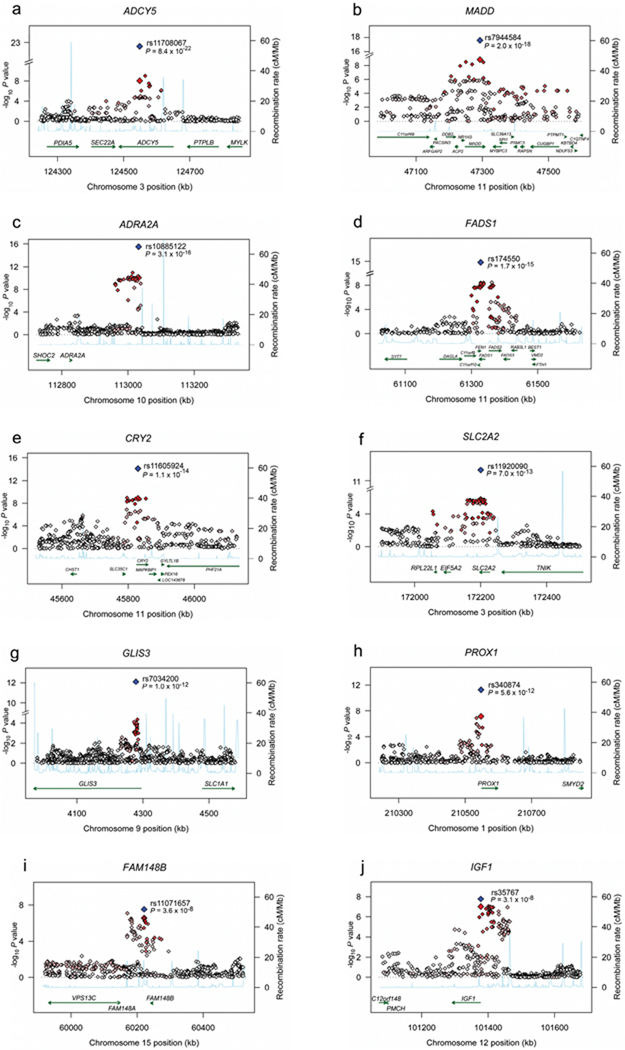
Figure 1.
Regional plots of ten novel genome-wide significant associations. For each of the ADCY5 (a), MADD (b), ADRA2A (c), FADS1 (d), CRY2 (e), SLC2A2 (f), GLIS3 (g), PROX1 (h), FAM148B (i) and IGF1 (j) regions, directly genotyped and imputed SNPs are plotted with their meta-analysis P values (as –log10 values) as a function of genomic position (NCBI Build 35). In each panel, the Stage 1 discovery SNP taken forward to Stage 2 replication is represented by a blue diamond (with global meta-analysis P value), with its Stage 1 discovery P value denoted by a red diamond. Estimated recombination rates (taken from HapMap) are plotted to reflect the local LD structure around the associated SNPs and their correlated proxies (according to a white to red scale from r2 = 0 to 1, based on pairwise r2 values from HapMap CEU). Gene annotations were taken from the University of California Santa Cruz genome browser.