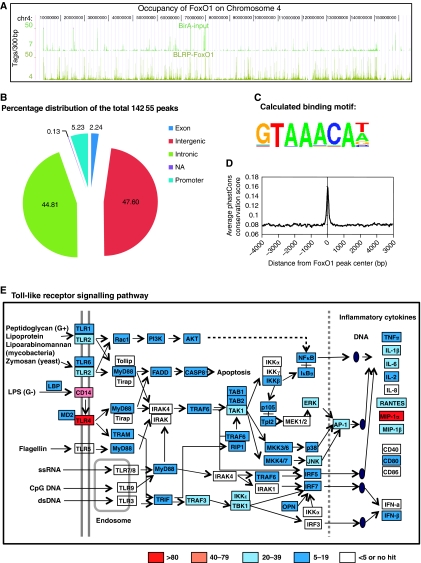
Figure 4.
Genome-wide localization of FoxO1 in RAW264.7 macrophage cells and FoxO1 occupancy on genes involved in Tlr signalling. (A) Browser tracks of BirA cells (blank control) and BirA–BLRP-FoxO1 ChIP-Seq peaks in chromosome 4. Note the high signal-to-noise ratios. (B–D) Location analysis of FoxO1-binding sites (peaks). (B) FoxO1-binding regions were mapped relative to their nearest RefSeq genes. The promoter region was defined as <1 kb segment upstream from the transcription start site (TSS). (C) Web logo of consensus motif position weight matricies (PWMs) generated by a de novo motif search of FoxO-binding sites. (D) Sequences of FoxO1-binding sites are highly conserved among different vertebrate species. (E) Gene Ontology analysis (using Kyoto Encyclopedia of Genes and Genomes pathways (KEGG)) of the genes located near FoxO1-binding regions (peaks) has identified Toll-like receptor signalling as one of the most-highly enriched pathways (P=2.62E−08; Bonferroni cut point: 8.47E−04). Genes have been coloured according to the intensity (height) of the dominant FoxO1 peak on each corresponding gene.