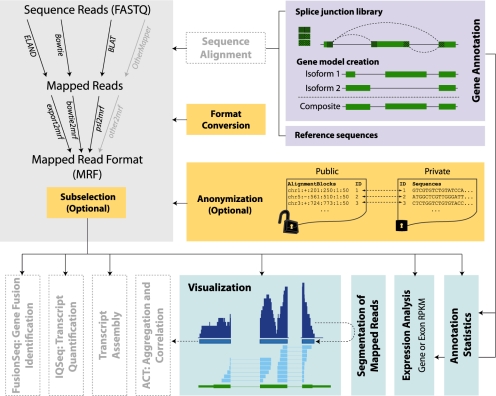
Fig. 1.
Schematic overview of RSEQtools. Mapped reads are first converted into MRF from common alignment tool output formats, including SAM. The resulting MRF files can be divided in two files: one with the alignment only and another with the corresponding sequence reads. The read identifiers provide a mapping between the two files. Then, several modules perform the downstream analyses independently from the mapping step, such as expression quantification, visualization of the mapped read and the calculation of annotation statistics, etc. Other tools have been developed based on this framework to perform more sophisticated analyses such as transcript assembly, isoform quantification (IQSeq, http://rnaseq.gersteinlab.org/IQSeq), fusion transcript identification (FusionSeq, http://rnaseq.gersteinlab.org/fusionseq), as well as aggregation and correlation of signal tracks (ACT, http://act.gersteinlab.org).