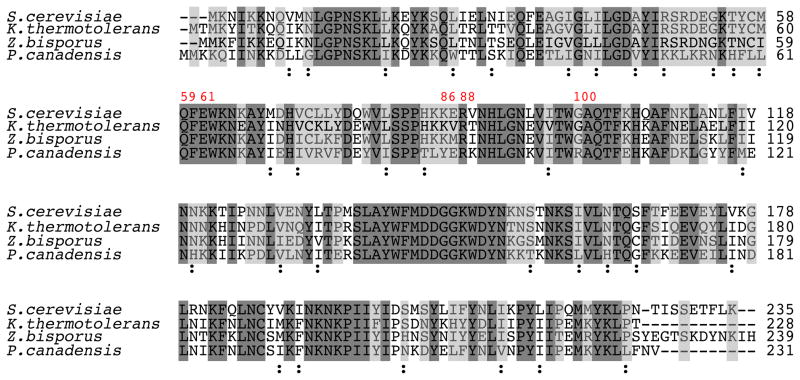
Figure 3.
Alignment of the amino acid sequence of I-SceI from S. cerevisiae with those of I-SceI homologues from Kluyveromyces thermotolerans, Zygosaccharomyces bisporus and Pichia canadensis. The positions of five residues randomized in the study are numbered in red. Residues identical in all homologues are highlighted in dark gray, and residues identical in I-SceI and in at least one other homologue are highlighted in light gray. Non-identical, conserved substitutions common in all four homologues are labeled with the symbol “:”. ClustalW2 was used to perform alignments and to define conserved substitutions55.