Figure 1.
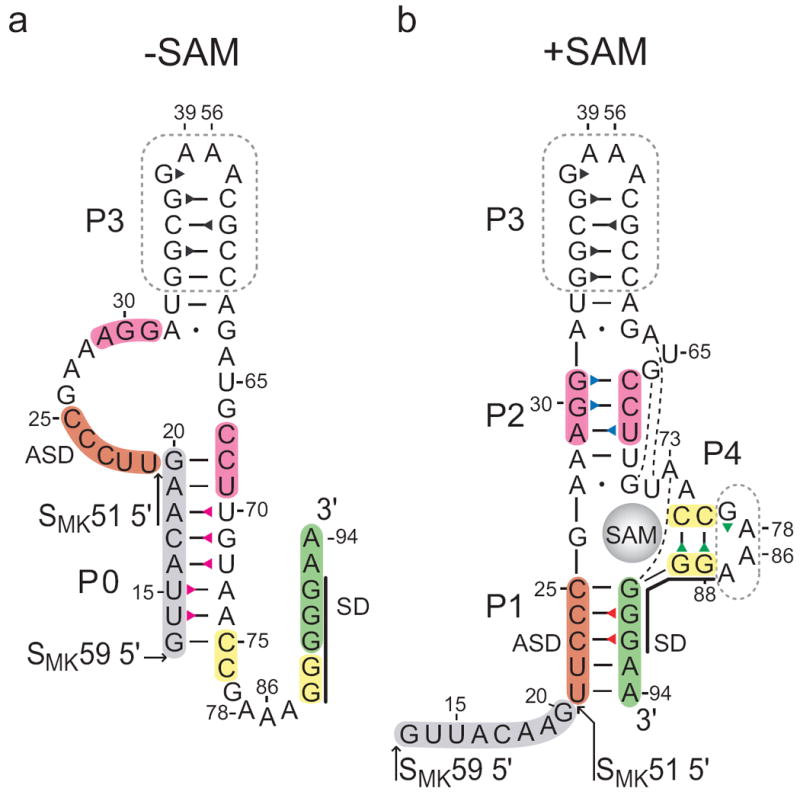
Secondary structure of the SMK box riboswitch. Previously proposed SAM-free (a) and SAM-bound (b) secondary structure models of the SMK box, represented as the SMK59 construct used in this study. The SMK51 construct lacks the eight 5’ nucleotides (grey). The boxed regions were truncated from hypervariable regions in the naturally-occurring E. faecalis metK sequence. Numbering is based on the E. faecalis metK sequence and is discontinuous at the engineered tetraloops. The Shine-Dalgarno (SD) sequence is marked with a black line. Base pairs observed in the crystal structure10 are marked: dash, Watson-Crick; dot, non-canonical; broken line, extrahelical. Triangles represent assigned imino protons and are colored corresponding to the labels in Figures 2 and 3.
