Figure 3.
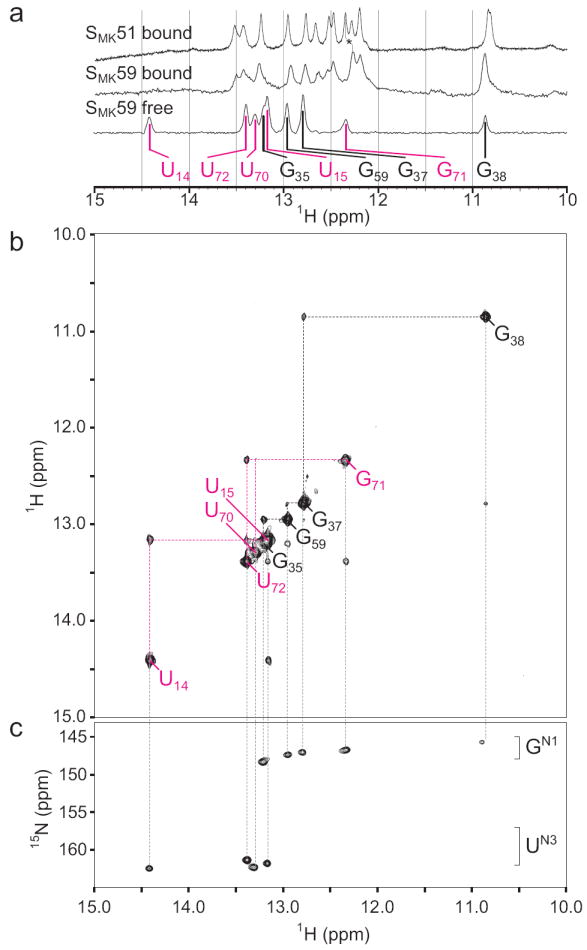
NMR spectra of SMK59 at 4°C reveal a novel fold in the absence of ligand. (a) The imino region of 1D 1H spectra of SAM-bound (top) SMK51, SAM-bound SMK59 (middle), or SAM-free SMK59 (bottom). Similarity of chemical shifts indicates near-identical structures for the SAM-bound constructs. A resonance shifted between the two bound states was used for assignment of G92 and is marked with an asterisk. The helix P3 spin network is labeled in black as in Figure 2; signals from the helix P0 are labeled in magenta (represented as color-coded triangles in Figure 1a). (b) Imino region of the 1H-1H NOESY spectrum of SAM-free SMK59. The helix P3 spin network is largely unperturbed compared to the SAM-bound state, but a novel spin network corresponding to helix P0 is detected. (c) 1H-15N HSQC spectrum of SAM-free SMK59 allows assignment of nucleotide type.
