Figure 5.
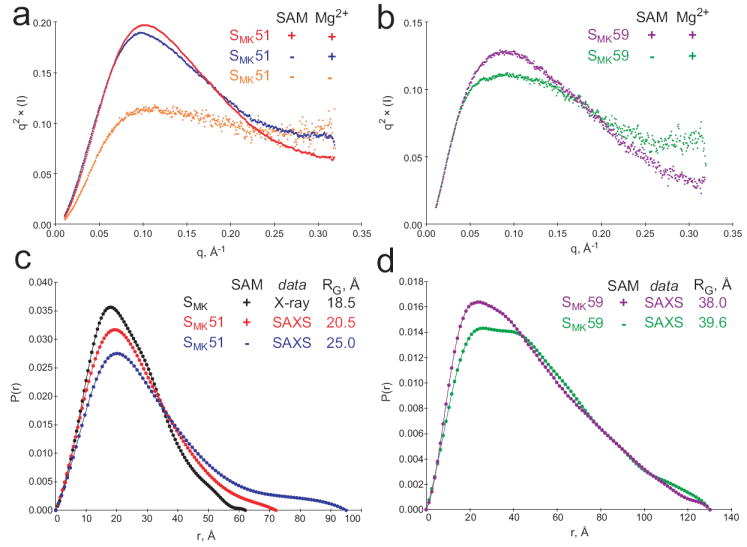
SAXS analysis of the SMK box in the presence and absence of ligand. (a) Kratky plots reveal increased folding of SMK51. Unfolded biopolymer is typified by a q2 × (I) plateau at high scattering angles (q > 0.2 Å-1), and this character decreases upon the addition of Mg2+ (blue) and SAM (red). (b) SAM-associated folding can also be observed in Kratky plots of SMK59 in the presence (purple) or absence (green) of SAM. (c) Intramolecular pair distribution functions for the SMK box. A simulated curve was generated for the previously-determined crystal structure of 53-nucleotide SMK box bound to SAM (black) and is compared to curves resulting from SAXS analysis of the 51-nucleotide SMK51 in solution (red, with SAM; blue, without SAM). Comparison of the radius of gyration (RG) suggests that the solution conformation is less compact than that adopted in the crystal. The binding of SAM is associated with a compaction of the solution structure of SMK51 based on the similar curve shapes and markedly different RG for free (25.01 Å, blue) versus bound (20.52 Å, red). (d) Comparison of SAXS-derived pair distribution curves for SMK59 with (purple) and without (green) SAM reveals a noteworthy change in the overall shape of the molecule. While the SAM-bound state generates a curve resembling those from the SAM-bound SMK51 species (c), the bimodal distribution in the absence of SAM is indicative of a two-lobed structure as might be expected for the ISOSMK state which is composed of two helical segments connected by a single-stranded region.
