Figure 6.
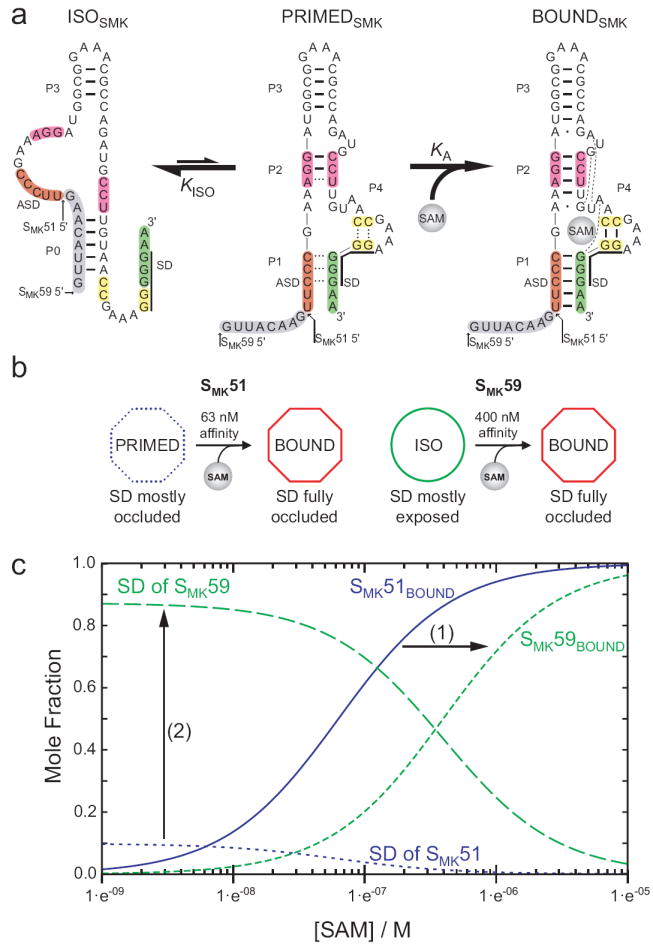
Model of SMK box structural transitions in response to SAM. (a) Observed conformations of SMK box. Left, the free state observed in SAM-free SMK59 via NMR, with accessible SD sequence. Center, the poorly-folded state observed via NMR in SAM-free SMK51 (which is missing the eight 5’ nucleotides shown here in grey), with a partially-accessible SD sequence. Right, the well-folded SAM-bound state with occluded SD sequence, observed in ligand-bound SMK51 and SMK59 and the previously-determined crystal structure. Hydrogen bonds are represented as lines as observed via NMR for ISOSMK and PRIMEDSMK, while hydrogen bonds expected based on the crystal structure10 are shown for BOUNDSMK. Dashed lines in the PRIMEDSMK state represent transiently formed base pairs as detected via NMR. (b) A schematic representing the transitions observed in ITC for SMK51 or SMK59 and how the analogous constructs behave while undergoing similar transitions inside the cell, as measured by β-galactosidase reporter assays. (c) Effect of the alternatively folded conformation on the populations of riboswitch conformations. SAM dependence of molecular species are simulated from equations (2, 3) describing the fractional populations of the SAM-bound species (SMK51BOUND, and SMK59BOUND) and of the effective concentration of exposed SD sequence for each construct. Curves were generated with the experimentally determined values of KA and KISO, and using a relative accessibility of the SD in the PRIMEDSMK form, α, estimated to be 0.1. The equilibrium between the ISOSMK and PRIMEDSMK conformations (1) shifts the affinity of the riboswitch into the biologically-relevant micromolar range, and (2) amplifies the response of the switch to SAM by promoting greater SD accessibility at low [SAM].
