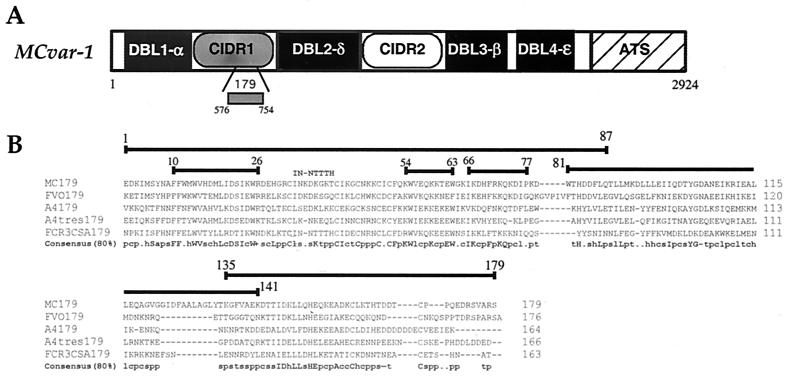
Figure 1.
Schematic representation and alignment of the amino acid sequences of the 179-aa minimal CD36-binding domain. (A) Schematic representation and amino acid boundaries of the MCvar1 gene (AAB60251) and the 179-aa CD36-binding domain. (B) Alignment of the amino acid sequences of the 179 region of PfEMP1 from various P. falciparum parasites. The consensus at 80% homology is also given. Alignment was performed by using the clustalw program (http://www.ebi.ac.uk/clustalw/), and the 80% consensus was performed by using the consensus program (http://www.bork.embl-heidelberg.de/cgi/consensus). Uppercase letters indicate conserved residues by the single-letter amino acid code. Lowercase letters indicate conserved classes of amino acids as follows: h, hydrophobic residues (A, C, F, G, H, I, K, L, M, T, T, V, W, Y); p, polar residues (C, D, E, H, K, N, Q, R, S, T); c, charged resides (D, E, H, K, R); a, aromatic residues (F, H, W, Y); s, small residues (A, C, D, G, N, P, S, T, V); +, positive residues (H, K, R); −, negative residues (D, E); l, aliphatic residues (I, L, V); and t, turnlike residues (A, C, D, E, G, H, K, N, Q, R, S, T). Peptides from the MC sequence used in various assays are indicated with their amino acid boundaries. The mutated site of the MC179 sequence (IN-NTTTH) also is indicated.