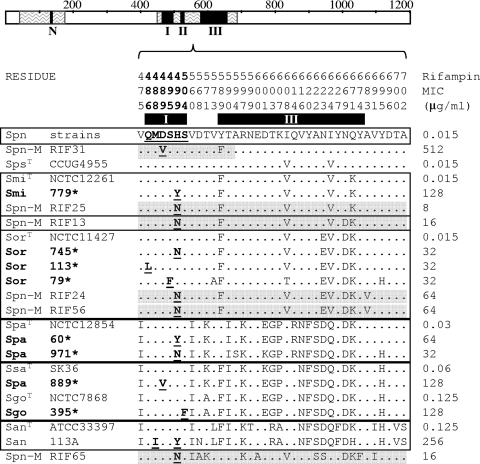
FIG. 2.
Amino acid sequence variations in RpoB (V475 to A702) of rifampin-resistant recombinant isolates of S. pneumoniae (Spn-M) and SMG rifampin-resistant isolates characterized in this work (boldface and marked with an asterisk). RpoB is represented as a bar with clusters N, I, II, and III as black boxes and zigzagged areas showing sequenced areas. The amino acids present at each polymorphic site are shown in full for S. pneumoniae strains (R6, P1031, Hungary, Taiwan1, TIGR4, and JJA). For the other strains, only sites that differ from those are shown. Residue numbers are indicated vertically above the sequences, and black boxes below the numbers localize clusters I and III. Amino acid changes involved in rifampin resistance are shown in boldface and underlined. Species nomenclature is as defined in the Fig. 1 legend. A superscript T indicates a type strain. Recombinant sequences are shadowed in gray. Squares group sequences with the highest similarity according to scores obtained by ClustalW alignments.