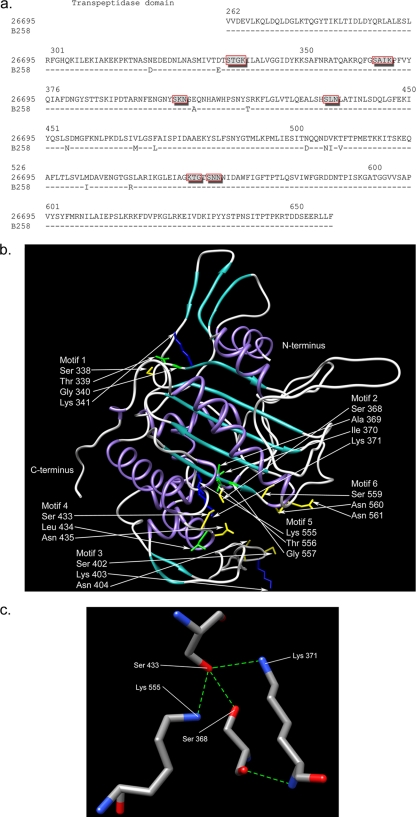
FIG. 1.
(a) Alignment of the PBP1 transpeptidase domains from 26695 and B258. The dashes indicate no change in B258 at these loci in comparison to the 26695 sequence. Specific amino acid substitutions are denoted by the letter of the changed amino acid. Boxed amino acids indicate putative penicillin binding motifs (PBMs). (b) Secondary structure of PBP1 transpeptidase region from 26695 depicted in ribbon representation (purple, alpha-helices; cyan, beta-sheets; gray, loops). The putative PBMs (boxed amino acids in panel a) are also shown, with amino acid side chains depicted in stick representation and colored by type (green, hydrophobic; yellow, polar; blue, polar positively charged). All other side chains have been deleted for clarity. (c) Hydrogen bond network within the putative binding cleft of the PBP1 transpeptidase region from 26695, involving the putative catalytic Ser 368 with Ser 433, Lys 371, and Lys 555 (blue, nitrogen; red, oxygen; grey, carbon; green dashed lines, hydrogen bonds). The putative catalytic Ser 368 is also amenable to weak hydrogen bonds with Lys 371 and Lys 555 (dashed lines not shown).