Abstract
Fungus-growing termites play an important role in lignocellulose degradation and carbon mineralization in tropical and subtropical regions, but the degradation potentiality of their gut microbiota has long been neglected. The high quality and quantity of intestinal microbial DNA are indispensable for exploring new cellulose genes from termites by function-based screening. Here, using a refined intestinal microbial DNA extraction method followed by multiple-displacement amplification (MDA), a fosmid library was constructed from the total microbial DNA isolated from the gut of a termite growing in fungi. Functional screening for endoglucanase, cellobiohydrolase, β-glucosidase, and xylanase resulted in 12 β-glucosidase-positive clones and one xylanase-positive clone. The sequencing result of the xylanase-positive clone revealed an 1,818-bp open reading frame (ORF) encoding a 64.5-kDa multidomain endo-1,4-β-xylanase, designated Xyl6E7, which consisted of an N-terminal GH11 family catalytic domain, a CBM_4_9 domain, and a Listeria-Bacteroides repeat domain. Xyl6E7 was a highly active, substrate-specific, and endo-acting alkaline xylanase with considerably wide pH tolerance and stability but extremely low thermostability.
The bioconversion of lignocellulosic biomass, the most abundant renewable resource on earth, contributes significantly to the production of alternative biofuels and organic chemicals (35). Therefore, increasing attention has been focused on exploring lignocellulase genes from diverse organisms, including free-living bacteria and fungi (45) as well as gut microbiomes of lignocellulose-consuming ruminants and insects (14, 18, 24, 46). Termites feature differentiated body plans, including masticating organs, enlarged hindguts, symbiotic systems, and enriched digestive enzymes, for thriving on complex diets consisting of different plant components in different termite species (2). Thus, termites are believed to possess diverse sets of efficient microscale lignocellulose bioconversion systems, and the exploration of this single community may reveal comprehensive hydrolytic enzymes responsible for the degradation of broad categories of plant polysaccharide bonds.
Termites of the subfamily Macrotermitinae, broadly known as termites that grow in fungi, are assumed to be one of the most abundant and influential insects in tropical and subtropical ecosystems in Asia and Africa (53). As a monophyletic lineage in the higher termites (33), members of this subfamily can specifically cultivate basidiomycete fungi of the genus Termitomyces in their nests. For most fungus-growing termites, the fungus comb is made from their primary feces, which are composed of partially digested dead plant materials (52). As the fungi grow, host termites gain nutrition by consuming the fungal nodules as well as the mature parts of the fungus comb (20). Therefore, fungus-growing termites actually develop a sophisticated triple symbiotic relationship with both their intestinal microbiota and ectosymbiotic fungi. Continuously in the past, Termitomyces spp. have been hypothetically assumed and experimentally proven to contribute diverse glycosyl hydrolases in plant material decomposition (22, 28, 29). Besides, some host termites have also been observed to secrete their own cellulases, including endo-1,4-β-glucanase and β-glucosidase, in their salivary glands or midgut (27, 44, 50). Thus, to date, little attention has been paid to the hydrolyzing capabilities of these intestinal microorganisms, except for fermentation and acetogenesis or methanogenesis (6). We believe it possible that intestinal microorganisms may also play a certain role in the overall process of lignocellulose digestion for their hosts.
Termite guts are special environmental ecosystems featuring steep pH, oxygen, and substrate gradients (8, 9), which are difficult to reproduce in vitro. As the majority of microorganisms in termite guts are unculturable, metagenomics becomes an ideal tool for exploring these presently inaccessible metagenome reservoirs (17), just as it has played significant roles in exploring many other environmental ecosystems (16, 38). However, the fact that microgram quantities of environmental DNA (eDNA) are required for the construction of screening or sequencing libraries has greatly limited its application, especially for low-biomass environments and rare samples, which potentially contain enzyme-encoding genes with great promise for biochemical, medical, and industrial applications (40). Fortunately, as a newly developed technique of whole-genome amplification, multiple-displacement amplification (MDA) has proven to be a powerful tool for overcoming this limitation in dealing with both individual uncultivated cells (25, 55) and whole-community metagenomes (1, 51).
Xylanases, a set of diverse enzymes such as endo-1,4-β-xylanase (EC 3.2.1.8), β-xylosidase (EC 3.2.1.37), α-arabinofuranosidase (EC 3.2.1.39), α-glucuronidase (EC 3.2.1.131), and acetylxylan esterase (EC 3.1.1.72), are responsible for the complete degradation of xylan, a major component of hemicelluloses. Among these, endo-1,4-β-xylanase is of greater importance, because it hydrolyzes the backbone of xylan into xylooligosaccharides, which is a critical step in the depolymerization process of xylan. Owing to this capability, endo-1,4-β-xylanase has played versatile roles in a variety of traditional industries, including pulp and paper, bioethanol, animal feed, textile, and waste treatment (41). Most importantly, as xylan is the second most abundant renewable organic carbon source on earth, its hydrolytic product, xylose, is inevitably an indispensable resource in the bioethanol conversion industry (19). Recent advancements concerning the manipulation of some bacteria, yeasts, and filamentous fungi to ferment pentose to ethanol (48) have highlighted its future applications. Additionally, the timely removal of hemicellulose backbones may enhance the hydrolytic efficiencies of cellulases by increasing their access to cellulose. All these properties indicate the great potential that xylanases hold to reduce the overall cost of ethanol production from lignocelluloses.
In the present study, considering the low DNA yield from the sampled termite intestinal metagenome and the potentiality of MDA, a fosmid library was constructed with MDA-amplified eDNA, allowing the isolation of 12 β-glucosidase-positive clones and one xylanase-positive clone. The xylanase gene was heterologously expressed, and the recombinant enzyme was biochemically characterized for its potential industrial applications.
MATERIALS AND METHODS
Chemicals and reagents.
The screening substrates carboxymethyl cellulose, 4-methylumbelliferyl-β-d-cellobioside (4-MUC), esculin hydrate, ferric ammonium citrate, and birchwood xylan were purchased from Sigma. Polysaccharide substrates (beechwood xylan, barley glucan, locust bean gum, starch, and Avicel) were purchased from Sigma. Restriction endonucleases, T4 DNA ligase, and Kod Plus DNA polymerase were obtained from Takara (Japan). The AxyPrep plasmid miniprep kit, AxyPrep DNA gel extraction kit, and AxyPrep PCR cleanup kit were purchased from Axygen. All other chemicals and reagents were from Sangon (People's Republic of China) and were of analytical grade.
Bacterial strains and plasmids.
EPI300-T1R (Epicentre) was used as the host strain for fosmid library cloning. Escherichia coli BL21 (Novagen) was used as the host strain for xylanase gene cloning and expression. The plasmids pCC2FOS (Epicentre) and pET-22b(+) (Novagen) were used as the fosmid vector and expression vector, respectively.
Termite collection and evisceration.
About 2,500 worker termites and 100 soldier termites closely related to Macrotermes annandalei (family Termitidae, subfamily Macrotermitinae) were collected from a nest in Xishuang Banna, Yunnan Province, People's Republic of China, in March 2008. After the termites were surface sterilized in 70% ethanol for 1 min and rinsed in phosphate-buffered saline (PBS) (8 g/liter NaCl, 0.2 g/liter KCl, 1.44 g/liter Na2HPO4, 0.24 g/liter KH2PO4 [pH 7.4]), the entire guts of the worker termites were immediately removed and transferred into 1.5-ml sterilized Eppendorf tubes containing 100 μl of PBS. The tubes, each containing about 100 guts, were set on ice. Additionally, about 100 intact soldier termites were collected in a tube for termite species identification. All samples were quickly frozen in liquid nitrogen and stored at −80°C until use. Besides morphological identification, the sampled termite was further identified with the molecular marker mitochondrial cytochrome oxidase subunit II (Co II) gene based on a method described previously by Ohkuma et al. (33).
Intestinal microbial DNA extraction.
Microbial DNA was extracted from termite guts using a modified indirect method, which can be roughly divided into three steps. First, to release the microbial cells from the termite gut, 1 ml (5 to 6 sample volumes) of trypsin-EDTA (PBS [pH 7.4] with 0.25% trypsin and 0.02% EDTA) was added to each tube of thawed sample, which contained about 100 μl of PBS with 100 termite guts, and the tubes were incubated at 37°C for 30 min with gentle inversion every 5 min to fully mix the gut tissue and enzyme. Trypsin digestion was terminated by the addition of 300 μl of 10% bovine serum albumin (BSA) (0.3 volumes of trypsin-EDTA), and a 1-ml pipette was used to homogenize the sample until no obvious gut particles could be observed. The samples were then vortexed for 30 s to ensure full homogenization.
Second, to isolate the microbial cells from the gut debris, the homogenized sample was centrifuged at 800 × g at 4°C for 10 min to remove the luminal debris, gut tissue, epithelial cells, and nuclei released from some of the lysed epithelial cells. The supernatant was gently and carefully collected and saved. The pellet was resuspended in 1 ml of PBS buffer and subjected to another two rounds of low-speed centrifugation as described above. The supernatants obtained from the three centrifugations were pooled and further clarified by two rounds of centrifugation at 800 × g, followed by centrifugation at 9,000 × g for 15 min at 4°C to collect the microbial cells.
Finally, the DNA was extracted according to a protocol described previously by Zhou et al. (57), with minor modifications. Briefly, the cell pellet was resuspended in 1 ml of PBS, and the following components were added: 668 μl of extraction buffer (100 mM Tris-HCl, 100 mM Na3PO4, 100 mM Na2EDTA, 1.5 M NaCl, 1% [wt/vol] cetyltrimethylammonium bromide [CTAB] [pH 8.0]), 100 μl of protease K (8 mg/ml in extraction buffer), and 32 μl of 25% SDS. The tube was incubated horizontally at 55°C for 20 min with gentle rotation for 10 min and then incubated at 70°C for 10 min. The sample was centrifuged at 17,000 × g for 10 min at 4°C. The supernatant was then used for phenol-chloroform extraction, and DNA was purified according to the protocol described previously by Zhou et al. (57).
Multiple-displacement amplification and evaluation.
The guts of the sampled termites were compacted with much soil and humic acid, and the DNA yields were rather low. To obtain enough total DNA for the construction of a fosmid library, the whole-community genomes were amplified by MDA with a REPLI-g minikit (Qiagen) according to the manufacturer's instructions. As amplification bias may be severe when the DNA template is less than 1 ng (55), we used a gradient of 10 to 75 ng DNA as templates of MDA to reduce the amplification bias. The amplified DNA was linearized by a three-step linearization process described by Zhang et al. (55), including phi-29 polymerase debranching, S1 nuclease digestion, and DNA polymerase I nick translation. To analyze the representational bias induced by MDA, the fingerprints of the V3 section of the 16S rRNA gene before and after MDA were compared. The amplification primers for the V3 section of the 16S rRNA gene were P2 (5′-ATTACCGCGGCTGCTGG-3′) and P3 (5′-GC CGC CCG CCG CGC GCG GCG GGC GGG GCG GGG GCA CGG GGG GCC TAC GGG AGG CAG CAG-3′). The PCR and amplification procedures were based on methods described previously by Ovreas et al. (34). Denaturing gradient gel electrophoresis (DGGE) was performed by using a DCode system (Bio-Rad Laboratories) according to standard procedures. In brief, about 2 μg of each PCR product was separated by electrophoresis in 8% polyacrylamide gels containing a 25 to 60% gradient of formamide and urea in 1× TAE (40 mM Tris, 1 mM Na2EDTA, 20 mM glacial acetate [pH 7.4]) for 5 h at 60°C and at a constant voltage of 200 V. After electrophoresis, the gel was silver stained (12) and digitally photographed.
Fosmid library construction and functional screening for cellulases and xylanase.
Gel purification, electroelution, and concentration of the MDA-amplified eDNA were performed by exactly following the protocols described previously by Brady (4). The whole-gut microbiome metagenome library was constructed with a CopyControl pCC2FOS fosmid library production kit (Epicentre), according to the manufacturer's instructions. Infected cells were grown on LB agar plates containing chloramphenicol (12.5 μg/ml) at 37°C for 14 h. A total of 10,000 transformants were selected and preserved in 384-well plates. With a 384-well inoculating needle holder, each 384-well plate was manually printed onto four plates with 12.5 μg/ml of chloramphenicol and different screening substrates, respectively. To screen for endoglucanase activity, the library clones were grown on LB agar plates containing 0.5% carboxymethyl cellulose for 12 h at 37°C, and the colonies were then stained with 0.2% Congo red for 30 min and destained with 1 M NaCl (42). The colonies surrounded by a yellow halo against a red background were identified as cellulase-positive clones. To screen for xylanase activity, the same procedure was performed, except that 0.5% birchwood xylan was used as a substrate. To screen for β-glucosidase activity, the library clones were grown on LB agar plates containing 0.1% esculin hydrate and 0.25% ferric ammonium citrate for 12 h at 37°C. Colonies with a black zone were selected as β-glucosidase-positive clones (26). To screen for cellobiohydrolase activity, library clones were first grown on LB agar plates for 12 h at 37°C and then overlaid with 0.1 M citrate-phosphate buffer (pH 7.0) containing 0.6% agarose and 0.04% 4-methylumbelliferyl-β-d-cellobioside (4-MUC), followed by further incubation for 1 h at 37°C. Colonies emitting fluorescence under UV light and lacking β-glucosidase activity were assumed to be candidate cellobiohydrolase-positive clones (36).
Sequencing and gene analysis of the xylanase-positive clone.
Whole-fosmid DNA from a mixed sample of all 13 positive clones was extracted by using a Large-Construction kit (Qiagen) according to the manufacturer's instructions. After 454 pyrosequencing (Roche Diagnostics) and assemblage, the xylanase-containing contig was analyzed in detail. Open reading frame (ORF) analysis was performed by using ORF Finder from the NCBI (http://www.ncbi.nlm.nih.gov/gorf/gorf.html). ORF sequence annotation was conducted by using BlastP and the NCBI nonredundant protein database (http://www.ncbi.nlm.nih.gov). For the xylanase-encoding ORF, the signal peptide was predicted by SignalP 3.0 in CBS (http://www.cbs.dtu.dk/services/SignalP/). Molecular mass and isoelectric point predictions were made with ExPASy (http://www.expasy.ch/tools/protparam.html). Multiple-sequence alignments were all conducted with ClustalW (43) and were based on the full length of selected sequences. Phylogenetic analysis was performed with MEGA 3.1, and an unrooted phylogenetic tree was generated by the neighbor-joining method.
Protein expression and purification.
The xylanase-encoding gene, designated xyl6E7, was amplified from the fosmid DNA by 35 cycles of PCR using forward primer 5′-ATCCCATGGTGGCACAAACAACTACTT-3′, which includes an NcoI restriction site at the 5′ end, and reverse primer 5′-CTTCTCGAGTTTGCTACAATCATCCAAG-3′, which includes an XhoI restriction site at the 5′ site. PCR fragments were purified with an AxyPrep PCR cleanup kit, excised with NcoI and XhoI, and ligated into the corresponding sites in pET-22b(+). The recombinant vectors were transformed into E. coli BL21. A single BL21 clone carrying pET 22b(+)-xyl6E7 was grown at 37°C in LB medium containing ampicillin (50 μg/ml) to an optical density of 0.6, expression was induced by the addition of 50 μM isopropyl-β-d-thiogalactopyranoside (IPTG), and the cells were incubated at 28°C for 16 h, with shaking at 120 rpm. The cells were harvested by centrifugation at 9,000 × g for 10 min and resuspended in lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole [pH 8.0]). Cells were lysed by using a combination of lysozyme and ultrasonication, as follows. Lysozyme was added at 1 mg per ml of cell solution, and the sample was incubated on ice for 30 min, after which it was sonicated for 10 min (5-s bursts with 10-s intervals) using a sonicator equipped with a microtip. The lysate was clarified by centrifugation at 10,000 × g for 20 min, and the supernatant was collected as the crude extract. The soluble recombinant Xyl6E7 with a 6×His tag was purified by using a Ni-nitrilotriacetic acid (NTA) column (Qiagen), which was equilibrated with lysis buffer, washed with buffer (50 mM NaH2PO4, 300 mM NaCl, 60 mM imidazole [pH 8.0]), and eluted with buffer (50 mM NaH2PO4, 300 mM NaCl, 250 mM imidazole [pH 8.0]). The fractions containing active xylanase were pooled, dialyzed against 20 mM NaH2PO4 (pH 7.4) using a Vivaspin 6 instrument (GE Healthcare), and stored at 4°C for use. The purity and molecular mass of Xyl6E7 were determined by SDS-PAGE, and the protein concentration was determined with a DC protein assay kit (Bio-Rad Laboratories).
Xylanase activity assay.
To characterize the properties of Xyl6E7, xylanase activity was assayed by measuring the amount of reducing sugar released from birchwood xylan after the addition of a dinitro-salicylic acid reagent (31). The standard assay mixture contained 0.05655 μg of the enzyme and 1% birchwood xylan in a final volume of 0.1 ml of 50 mM NaH2PO4-Na2HPO4 (pH 7.5) and was incubated at 55°C for 10 min, in triplicate. The reaction was terminated by the addition of 100 μl of 3,5-dinitrosalicylic acid (DNS) and boiling for 5 min. The absorbance at 540 nm was measured by using a Multiskan Spectrum spectrophotometer (Thermo Scientific, Finland). Xylanase activity (U/mg) was defined as micromoles of reducing sugar released from xylan per minute per milligram of enzyme protein under the standard assay conditions stated above. To test the pH range of Xyl6E7, a standard activity assay was performed with 50 mM appropriate buffers between pH 4 and 10.5: sodium-acetate buffer was used for pH 4 to 5.5, sodium phosphate buffer was used for pH 6 to 8.5, and N-cyclohexyl-3-aminopropanesulfonic acid was used for pH 9.0 to 10.5. The pH stability was determined by measuring the residual activity after the preincubation of the enzyme in the buffers between pH 4.5 and 10.0 at 4°C for up to 6 days. To determine the temperature range over which the enzyme was active, the standard activity assay was performed at between 20°C and 70°C. Thermostability was determined by measuring the residual activity after the preincubation of the enzyme for 5, 15, and 30 min at each of the temperatures 30°C, 40°C, 45°C, 50°C, and 55°C. The protective effect of l-cysteine (50 mM) was also tested. To test the substrate specificity of Xyl6E7, birchwood xylan was replaced in the standard assay by beechwood xylan, barley glucan, locust bean gum, starch, and Avicel. The Xyl6E7 activity was also measured by using the standard assay after incubating the enzyme with various chemicals and metal ions at 1 mM for 30 min at 4°C to assess their effects. For kinetic analysis, the activity of Xyl6E7 was measured with birchwood xylan at concentrations ranging from 0.2 to 11 mg/ml, and the Km and Vmax were calculated according to the Lineweaver-Burk method.
Thin-layer chromatography of hydrolysis products.
To investigate the reaction pattern of Xyl6E7, the hydrolysis products of birchwood xylan for 10 min with 0.03665 U of Xyl6E7 and for 12 h with 3.665 U of Xyl6E7 were analyzed by thin-layer chromatography (TLC). The products were spotted onto a silica gel 60 F254 plate and developed at room temperature with n-butanol-formic acid-water (4:6:1, vol/vol/vol) as a spray reagent. The spots were visualized by spraying the plates with a chromogenic solution containing 20 mg of cerous sulfate, 50 mg of ammonium molybdate, 5 ml of H2SO4 (98%), and 95 ml of ethanol (56), followed by incubation at 121°C for 5 min afterwards. Xylose, xylobiose, and xylotriose were used as standards.
Nucleotide sequence accession numbers.
The GenBank accession no. for xyl6E7 is HM483387, and that for the Co II gene of Macrotermes annandalei is HM483386.
RESULTS
Termite species identification, DNA extraction, MDA, and validation.
The sequence of the mitochondrial cytochrome oxidase subunit II gene from the heads of 10 soldier termites showed maximum identity (96%) with the gene from M. annandalei (GenBank accession no. AB300696). Thus, the collected termites were classified as Macrotermes annandalei, subfamily Macrotermitinae, family Termitidae. As the guts of M. annandalei termites contained a large proportion of soil and humic acid, approximately 450 ng of DNA was obtained from about 100 M. annandalei worker termite guts, compared to several micrograms of DNA from equal numbers of other termite guts. The refined extraction method used in this study limited the contamination from termites to the total microbial DNA (eukaryotic contamination was estimated to be less than 1%, based on 454 pyrosequencing data for eDNA from termite species [data not shown]). For MDA, 10, 20, 37.5, and 75 ng of primary eDNA were used as templates, and all showed relatively equal amplification efficiencies, with yields of 2 μg. The highly similar DGGE fingerprints of the V3 regions of the 16S rRNA genes before and after MDA indicated that no obvious amplification bias could be identified by DGGE (see Fig. S1 in the supplemental material).
Functional screening for lignocellulases and sequence analysis of the xylanase gene.
All 10,000 fosmid clones from the intestine metagenomic library from fungus-growing termites were subjected to functional screening for four lignocellulose-degrading enzymes. In total, one xylanase-positive clone and 12 β-glucosidase-positive clones were identified. No clones positive for endoglucanase or cellobiohydrolase were identified.
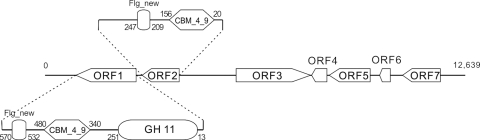
After sequencing and assembly, the xylanase-positive clone revealed a 12,639-bp contig containing 7 putative ORFs (Table 1), among which an endo-1,4-β-xylanase ORF and an adjacent CBM_4_9 ORF were obviously relevant to lignocellulose degradation. The singular arrangement of the 7 ORFs on the contig can be seen in Fig. 1. The endo-1,4-β-xylanase ORF (ORF1) begins with the putative initiation codon ATG and ends with TAG. It consists of 1,818 nucleotides encoding a protein of 605 amino acids with a deduced molecular mass of 64,532 Da. No coding sequence for an apparent signal peptide was identified. Homology searches revealed that Xyl6E7 was a unique modular xylanase. Amino acids 13 to 251 exhibited significant homology to GH11 family catalytic modules and had the highest identity of 73% with that reported under GenBank accession no. AAS85783 (7) (a xylanase isolated from the gut of a lepidopteran caterpillar). Amino acids 340 to 480 showed significant homology to the CBM_4_9 module and had a maximal identity of 38% with that reported under accession no. ADD82900 (a xylanase isolated from termite gut). No obvious linker sequence could be detected between the two domains. Amino acids 532 to 570 showed significant homology to the Listeria-Bacteroides repeat domain, which belongs to the Flg_new superfamily and had the highest identity of 38% with that of a whole-genome-sequenced clostridial species, Coprococcus eutactus ATCC 27759 (accession no. EDP26952). ORF2 consisted of a CBM_4_9 module, which has a maximal identity of 45% with the carbohydrate-binding CenC domain protein from Fibrobacter succinogenes subsp. succinogenes S85 (accession no. ACX75364), and a Listeria-Bacteroides repeat domain, which had the highest identities of 50% with that from Mollicutes bacterium D7 (accession no. EEO33990). The similarity between the two CBM_4_9 modules on the fosmid contig was 43.8%, and that between the two Listeria-Bacteroides repeat domains was 76.9%.
TABLE 1.
Characteristics of fosmid fragments containing xyl6E7
| ORF | ORF position (range) | Length (aa)a | G+C content (%) | Best hit (GenBank accession no.) | Organism | % identity/% similarity | E value |
|---|---|---|---|---|---|---|---|
| 1 | 922-2739 | 605 | 44.9 | Endo-1,4-beta-xylanase precursor (AAS85783) | Uncultured bacterium | 73/82 | 4e−102 |
| 2 | 2904-4025 | 373 | 44.6 | Carbohydrate binding CenC domain protein, CBM_4_9 superfamily (ACX75364) | Fibrobacter succinogenes subsp. succinogenes S85 | 45/64 | 4e−28 |
| 3 | 5722-7965 | 747 | 43.4 | Guanosine-3′,5′-bis(diphosphate) 3′-pyrophosphohydrolase (ABR44269) | Parabacteroides distasonis ATCC 8503 | 61/79 | 0.0 |
| 4 | 7987-8451 | 154 | 43.9 | Acetyltransferase (ABR44010) | Parabacteroides distasonis ATCC 8503 | 73/86 | 8.6e−61 |
| 5 | 8536-9774 | 412 | 41.6 | Alkyl hydroperoxide reductase/thiol-specific antioxidant/malallergen (ACU58361) | Chitinophaga pinensis DSM 2588 | 30/51 | 3e−51 |
| 6 | 10019-10345 | 108 | 41.9 | Hypothetical protein PRABACTJOHN_02858 (ribosome-associated heat shock protein) (EEC95765) | Parabacteroides johnsonii DSM 18315 | 50/73 | 2e−25 |
| 7 | 10703-11842 | 379 | 43.1 | Thiol protease/hemagglutinin PrtT (EAY25069) | Microscilla marina ATCC 23134 | 29/49 | 5e−06 |
aa, amino acids.
FIG. 1.
ORF arrangement on the contig of the xylanase-positive clone. The full length of the xylanase-localizing contig is 12,639 bp, and it consists of 7 ORFs. The transcriptional direction of each ORF is indicated by the arrow orientation. Modular elements of ORF1 and ORF2, which are relevant to lignocellulose degradation, are shown in detail. In ORF1, residues 13 to 251 belong to the GH11 family catalytic modules, residues 340 to 480 belong to CBM_4_9 modules, and residues 532 to 570 show the highest similarity to the Listeria-Bacteroides repeat domain, which belongs to the Flg_new superfamily. In ORF2, residues 20 to 156 belong to CBM_4_9 modules, and residues 209 to 247 belong to the Listeria-Bacteroides repeat domain.
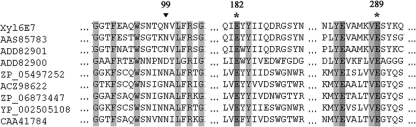
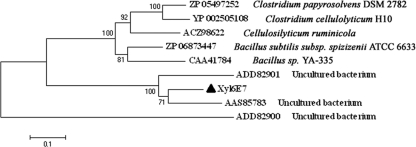
A BlastP search of the NCBI nonredundant protein database revealed that Xyl6E7 was closely related to eight putative GH11 xylanases (Fig. 2), the top three of which were all from xylophagous insect guts. Multiple-sequence alignments of these sequences revealed two conserved glutamate residues (positions 182 and 289), which are required for the catalytic activity of all GH11 xylanases (47), and one specific residue (position 99) that is well known to be responsible for the pH dependence property of most GH11 xylanases (37). This residue is located near the first catalytic glutamate residue and can be either an Asp (D) in those with an “acidic” pH optimum or an Asn (N) in more “alkaline” xylanases (23). The existence of an Asn (N) at residue 99 of Xyl6E7 indicated its potential alkaline property, which was supported by subsequent biological characterizations. Moreover, the proteins reported under GenBank accession no. AAS85783 (7), ACZ98622 (10), and CAA41784 (54), which also have Asn (N) at position 99, are also alkaline xylanases, further corroborating the influence of this residue on the GH11 xylanase pH profile. (Data for accession no. ADD82901, ADD82900, ZP_05497252, ZP_06873447, and YP_002505108 are all unpublished, so their pH profiles could not be obtained as yet.) Phylogenetic analysis of Xyl6E7 revealed that enzymes from similar insect origins are not necessarily clustered. Actually, Xyl6E7 was most closely related to the protein reported under accession no. AAS85783 (from caterpillar gut), while it was least closely related to that reported under accession no. ADD82900 (from termite gut) (Fig. 3).
FIG. 2.
Alignment of Xyl6E7 with eight of its most related GH11 xylanases. The protein reported under GenBank accession no. AAS85783 was derived from caterpillar gut, those under accession no. ADD82901 and ADD82900 were derived from termite gut, that under accession no. ZP_05497252 was from Clostridium papyrosolvens, that under accession no. ACZ98622 was from Cellulosilyticum ruminicola, that under accession no. ZP_06873447 was from Bacillus subtilis subsp. spizizenii ATCC 6633, that under accession no. YP_002505108 was from Clostridium cellulolyticum H10, and that under accession no. CAA41784 was from Bacillus sp. strain YA-335. Multiple-sequence alignments were performed with ClustalW. Identical residues are shaded in black. The two highly conserved catalytically essential glutamic acid residues of GH11 xylanases are marked by asterisks, and the residue known to be responsible for the pH dependence of GH11 xylanases is marked by a triangle.
FIG. 3.
Phylogenetic analysis of Xyl6E7. Multiple-sequence alignments were performed against the full length of Xyl6E7 and its eight closest GH11 xylanases (the same sequences as those shown in Fig. 2). An unrooted phylogenetic tree was then established with MEGA 3.1 by the neighbor-joining method, with bootstrap values for 1,000 resamplings shown at major nodes. GenBank accession numbers and organisms are shown for each sequence.
Protein expression, purification, and characterization.
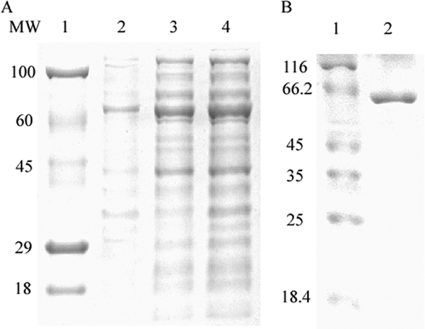
The endo-1,4-β-xylanase gene xyl6E7 was cloned into pET 22b(+) and expressed in E. coli BL21. The majority of the expressed Xyl6E7 was detected to be soluble in the supernatant of lysed cells (Fig. 4 A). With an N-terminal 6×His tag, Xyl6E7 was successfully purified on a Ni-NTA column (Qiagen). The size of the purified Xyl6E7 determined by SDS-PAGE was in good agreement with the predicted value (Fig. 4B). A substrate specificity assay revealed that Xyl6E7 had high levels of activity of 733 ± 20.6 U/mg toward birchwood xylan and 771 ± 17.2 U/mg toward beechwood xylan under optimal conditions but had no detectable activity against carboxymethyl cellulose, Avicel, starch, barley glucan, or locust bean gum. This high level of substrate specificity is a typical property of GH11 xylanases.
FIG. 4.
SDS-PAGE (12%) analysis of the purified xylanase. (A) Lanes: 1, stained protein molecular weight (MW) marker (in thousands); 2, insoluble protein fractions expressed by the induced bacteria; 3, soluble protein fractions expressed by the induced bacteria; 4, total proteins expressed by the induced bacteria. (B) Lanes: 1, unstained protein molecular weight marker; 2, purified Xyl6E7 (about 64.5 kDa).
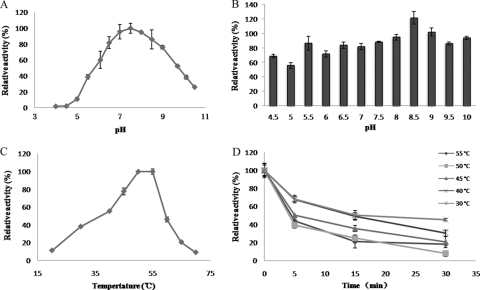
The impacts of pH and temperature on the activity and stability of Xyl6E7 were examined. The enzyme displayed a pH optimum of 7.5 and retained more than 50% of the maximal activity across a broad pH range of 6.0 to 9.5 (Fig. 5 A). Xyl6E7 was sensitive to low pH, exhibiting less than 10% of the maximal activity below pH 6.0. Xyl6E7 was very stable over a wide pH range, with more than 70% of residual activity after incubation at pH 5.5 to 10 for 6 days at 4°C (Fig. 5B).
FIG. 5.
Effects of pH and temperature on the activity and stability of Xyl6E7. (A) The pH range of Xyl6E7 was assayed at 55°C between pH 4.0 and 10.5. (B) pH stability was measured after preincubation in buffers ranging from pH 4.5 to 10 at 4°C for 6 days. (C) The temperature range was assayed between 20°C and 70°C at pH 7.5. (D) Thermostability was detected by measuring the residual activity after preincubation at 30°C to 50°C for 5 min, 10 min, and 30 min. All relative activity values were obtained from triplicate experiments. For more details, refer to Materials and Methods.
Xyl6E7 exhibited maximal activities between 50°C and 55°C and retained more than 40% of the maximal activity between 30°C and 60°C (Fig. 5C). A thermostability assay revealed that Xyl6E7 was more rapidly deactivated as the temperature increased and more greatly deactivated as the incubation time was extended. As shown in Fig. 5D, the activity of Xyl6E7 decreased to less than 70% of the initial activity after a 5-min preincubation at 30°C to 55°C, decreased to less than 50% after a 15-min incubation, and decreased further at the time point of 30 min. l-Cysteine (50 mM) was found to be able to restore the activity of Xyl6E7 from under 20% at 55°C for 30 min to over 60% under the same conditions, which may be attributable to the protection effect of disulfide bonds against reduction.
The substrate specificity revealed no detectable activity of Xyl6E7 toward Avicel, starch, locust bean gum, or carboxymethyl cellulose, which is in good accordance with the high level of substrate specificity of most GH11 xylanases. The effects of different metal ions and chemical reagents at 1 mM on Xyl6E7 are shown in Table 2. Taking the activity with no additional reagents as 100%, Xyl6E7 was almost completely inhibited by Ag+, Cu2+, and Fe3+; was significantly inhibited by Al3+, Zn2+, and Mn2+; and was partially inhibited by Ni2+, Ba2+, Co2+, EDTA, Ca2+, and K+ in the order Ni2+ > Ba2+ > Co2+ > EDTA > Ca2+ > K+. Tris-Cl and Mg2+ led to obvious increases in xylanase activity. The Vmax and Km of Xyl6E7 were determined based on a Lineweaver-Burk plot, which were 1,057.80 μmol min−1 mg−1 protein and 6.96 mg ml−1, respectively.
TABLE 2.
Effects of different metal ions and reagents on Xyl6E7 activity
| Chemical | Mean relative activity (%) at 1 mM ± SDa |
|---|---|
| Control | 100.00 ± 2.69 |
| Tris-Cl | 112.88 ± 2.53 |
| Ag+ | 6.80 ± 0.40 |
| Co2+ | 70.91 ± 4.02 |
| Cu2+ | 8.70 ± 0.72 |
| Fe3+ | 12.74 ± 2.94 |
| Ca2+ | 84.63 ± 3.67 |
| Mg2+ | 119.64 ± 1.83 |
| Mn2+ | 45.30 ± 4.95 |
| K+ | 87.92 ± 1.34 |
| Ba2+ | 67.89 ± 2.10 |
| Ni2+ | 65.49 ± 1.27 |
| Zn2+ | 40.99 ± 1.52 |
| Al3+ | 28.58 ± 0.94 |
| Lys | 100.47 ± 3.55 |
| EDTA | 79.12 ± 3.67 |
Relative activity values were obtained from three repeated experiments.
Thin-layer chromatography of hydrolysis products.
The primary and final hydrolysis products of Xyl6E7 on birchwood xylan were analyzed by TLC. The primary reaction products (0.03665 U for 10 min) were a series of short-chain xylooligosaccharides, including only a small fraction of xylotriose, while the final products (3.665 U for 12 h) were mainly xylotetrose, xylotriose, and xylobiose, with no appearance of xylose (Fig. 6). These results indicate that the purified enzyme was an endo-type xylanase.
FIG. 6.
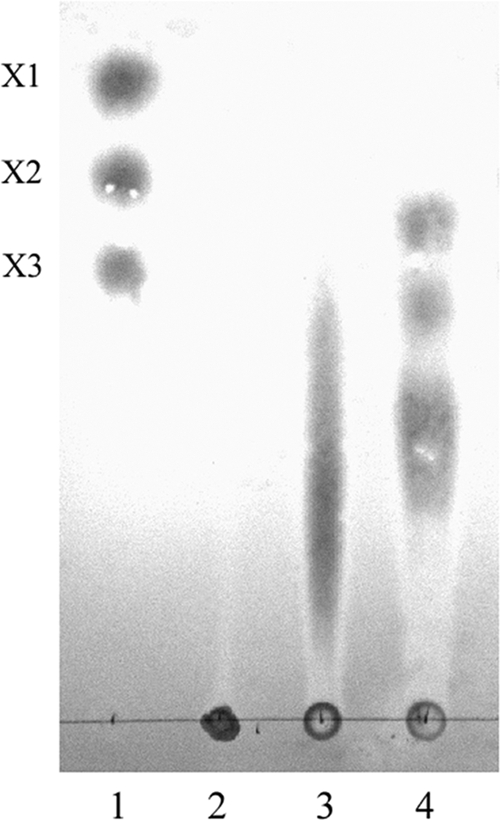
Thin-layer chromatography of the hydrolytic products of Xyl6E7. Lanes: 1, xylose (X1), xylobiose (X2), and xylotriose (X3) as standards; 2, birchwood xylan; 3, primary hydrolytic products (0.03665 U for 10 min); 4, final hydrolytic products (3.665 U for 12 h).
DISCUSSION
In this study, a metagenomic library was constructed from the guts of fungus-growing termites, and the process may contain helpful technical insights for similar studies. First, the whole gut, instead of only the luminal contents (49), was used as the initial sample; thus, members attached to the host intestinal epithelium were included to fully capture the intact metagenome. Second, a refined trypsin lysis procedure was adopted, instead of the commonly used grinding method (7) or the direct bead-beating method (30), to minimized the mechanical damage to intestinal epithelial cells. This step matters because nuclei released from damaged cells would be more difficult to extrude than intact epithelial cells. Modified differential centrifugation (800 × g compared to 1,000 × g [15]) was then used to eliminate intestinal debris, disassociated gut epithelium cells, and nuclei released from lysed cells, thus optimizing the purity and recovery of further DNA extraction and reducing host eukaryotic contamination. Third, a method of whole-genome amplification, MDA, was proactively and successfully applied as a powerful tool for obtaining sufficient quantities of eDNA for fosmid library construction and functional screening, despite the case revealed by some studies that chimeric sequences may still exist at the ends of fosmid contigs (11, 32). This method may prove efficient and useful for the functional screening of other low-yield environmental niches.
Thus far, the microbiome of fungus-growing termites has long been neglected for its hydrolytic potential, and the isolation of a batch of lignocellulase-positive clones directly demonstrates that it does contribute glycosyl hydrolases for their hosts' digestion. This finding expands gut microbes' routine roles of acetogenesis or methanogenesis and gives a new vision of the symbiosis system of fungus-growing termites. Besides, the preferential enrichment of β-glucosidases may indicate that gut microbes are especially good at hydrolyzing cellobiose to glucose monomers, although more-comprehensive high-throughput genomic information would be required to further explore the enzyme profile inherent in the gut microbiota of fungus-growing termites.
The phylogenetic analysis of Xyl6E7 and its related GH11 xylanases unexpectedly revealed that these enzymes were not simply clustered by their host origin. Actually, xylanases of termite origin (Xyl6E7 and those reported under GenBank accession no. ADD82900 and ADD82901) did not all fall into the same cluster. This may be attributable to differences of termite species feeding on different diets, which would inevitably feature different microbial community structures (5). The fosmid contig containing Xyl6E7 showed an interesting arrangement. It contained two sets of the CBM_4_9 domain flanked by a Listeria-Bacteroides repeat domain within and adjacent to Xyl6E7, respectively. As the CBM_4_9 family includes diverse carbohydrate binding domains, and most CBM modules are believed to be able to enhance the enzyme efficiency by increasing the accessibility of susceptible substrates to the enzyme catalytic module. Thus, the binding capability and specificity of CBM_4_9 inside and alongside Xyl6E7 are worthy of further investigation.
Compared to other xylanases of a metagenomic origin, a most remarkable advantage of Xyl6E7 may be its high efficiency, 733 U/mg against birchwood xylan and 771 U/mg against beechwood xylan at the defined optimal conditions, which placed it among the top natural bacterial xylanases documented in the BRENDA database (5,890 U/mg for Bacillus subtilis [3], 1,780 U/mg for Bacillus pumilus [33a], 867 U/mg for Thermotoga maritima [21], and 720 U/mg for Bacillus licheniformis [13]). Xyl6E7 is also impressive for its wide pH tolerance and stability, which would undoubtedly be beneficial for challenging pH processes or where neutralizing pH is inefficient or uneconomical. Unfortunately, in common with most members of GH11 xylanases, Xyl6E7 exhibits obvious low thermostability, which means that further enzyme-engineering efforts would be required for industrial conditions demanding high temperatures. Furthermore, the final hydrolysis products of Xyl6E7 are a series of xylooligosaccharides, which would have potential applications in food, feed formulation, and pharmaceutical industries.
In conclusion, we describe an optimized, yet simple, protocol for the preparation of metagenomic DNA from intestinal samples. This protocol can maximize the diversity of microbes represented and minimize host eukaryotic contamination without depending on large-scale devices or extravagant extraction kits, thereby allowing the optimal exploitation of the genetic potential of intestinal samples. Moreover, the successful combination of MDA and functional metagenomics has expanded the role of MDA as a powerful tool for library construction, providing access to the genomic reservoirs of niches with low DNA yields. We believe that this encouraging example can be extended to the further functional screening of DNA libraries from other promising, but limited, environmental habitats. More noticeably, the fact that the lignocellulase genes were reserved in the microbiome of fungus-growing termites revealed the neglected contributions of intestinal microbes to plant material digestion for host nutrition. Furthermore, we discovered a highly active xylanase with wide pH tolerance and stability, which are ideal properties for potential industrial applications.
Supplementary Material
Acknowledgments
This work was financially supported by the Knowledge Innovation Program of the Chinese Academy of Sciences (grants KSCX2-YW-G-062 and KSCX2-YW-G-022), the National High Technology Research and Development Program of China (863 Program) (grant 2007AA021302), the 100 Talents Program of the Chinese Academy of Sciences, the National Natural Science Foundation of China (grant 30900150), the Natural Science Foundation of Shanghai Province (grant 09ZR1436900), and the Cooperation Program between Guangdong Province and Chinese Academy of Science (grant 2009B091300147).
Footnotes
Published ahead of print on 5 November 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Abulencia, C. B., D. L. Wyborski, J. A. Garcia, M. Podar, W. Q. Chen, S. H. Chang, H. W. Chang, D. Watson, E. L. Brodie, T. C. Hazen, and M. Keller. 2006. Environmental whole-genome amplification to access microbial populations in contaminated sediments. Appl. Environ. Microbiol. 72:3291-3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bignell, D. E., P. Eggleton, L. Nunes, and K. L. Thomas. 1997. Termites as mediators of carbon fluxes in tropical forests: budgets for carbon dioxide and methane emissions, p. 109-134. In A. D. Watt, N. E. Stork, and M. D. Hunter (ed.), Forests and insects. Chapman and Hall, London, United Kingdom.
- 3.Bourgois, T. M., D. V. Nguyen, S. Sansen, S. Rombouts, T. Belien, K. Fierens, G. Raedschelders, A. Rabijns, C. M. Courtin, J. A. Delcour, S. Van Campenhout, and G. Volckaert. 2007. Targeted molecular engineering of a family 11 endoxylanase to decrease its sensitivity towards Triticum aestivum endoxylanase inhibitor types. J. Biotechnol. 130:95-105. [DOI] [PubMed] [Google Scholar]
- 4.Brady, S. F. 2007. Construction of soil environmental DNA cosmid libraries and screening for clones that produce biologically active small molecules. Nat. Protoc. 2:1297-1305. [DOI] [PubMed] [Google Scholar]
- 5.Brauman, A., J. Dore, P. Eggleton, D. Bignell, J. A. Breznak, and M. D. Kane. 2001. Molecular phylogenetic profiling of prokaryotic communities in guts of termites with different feeding habits. FEMS Microbiol. Ecol. 35:27-36. [DOI] [PubMed] [Google Scholar]
- 6.Brauman, A., M. D. Kane, M. Labat, and J. A. Breznak. 1992. Genesis of acetate and methane by gut bacteria of nutritionally diverse termites. Science 257:1384-1387. [DOI] [PubMed] [Google Scholar]
- 7.Brennan, Y., W. N. Callen, L. Christoffersen, P. Dupree, F. Goubet, S. Healey, M. Hernandez, M. Keller, K. Li, N. Palackal, A. Sittenfeld, G. Tamayo, S. Wells, G. P. Hazlewood, E. J. Mathur, J. M. Short, D. E. Robertson, and B. A. Steer. 2004. Unusual microbial xylanases from insect guts. Appl. Environ. Microbiol. 70:3609-3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brune, A. 1998. Termite guts: the world's smallest bioreactors. Trends Biotechnol. 16:16-21. [Google Scholar]
- 9.Brune, A., and M. Friedrich. 2000. Microecology of the termite gut: structure and function on a microscale. Curr. Opin. Microbiol. 3:263-269. [DOI] [PubMed] [Google Scholar]
- 10.Cai, S. C., J. B. Li, F. Z. Hu, K. G. Zhang, Y. M. Luo, B. Janto, R. Boissy, G. Ehrlich, and X. Z. Dong. 2010. Cellulosilyticum ruminicola, a newly described rumen bacterium that possesses redundant fibrolytic-protein-encoding genes and degrades lignocellulose with multiple carbohydrate-borne fibrolytic enzymes. Appl. Environ. Microbiol. 76:3818-3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, Y., M. G. Dumont, J. D. Neufeld, L. Bodrossy, N. Stralis-Pavese, N. P. McNamara, N. Ostle, M. J. I. Briones, and J. C. Murrell. 2008. Revealing the uncultivated majority: combining DNA stable-isotope probing, multiple displacement amplification and metagenomic analyses of uncultivated Methylocystis in acidic peatlands. Environ. Microbiol. 10:2609-2622. [DOI] [PubMed] [Google Scholar]
- 12.Creste, S., A. T. Neto, and A. Figueira. 2001. Detection of single sequence repeat polymorphisms in denaturing polyacrylamide sequencing gels by silver staining. Plant Mol. Biol. Rep. 19:299-306. [Google Scholar]
- 13.Damiano, V. B., R. Ward, E. Gomes, H. F. Alves-Prado, and R. Da Silva. 2006. Purification and characterization of two xylanases from alkalophilic and thermophilic Bacillus licheniformis 77-2. Appl. Biochem. Biotechnol. 129:289-302. [DOI] [PubMed] [Google Scholar]
- 14.Duan, C. J., L. Xian, G. C. Zhao, Y. Feng, H. Pang, X. L. Bai, J. L. Tang, Q. S. Ma, and J. X. Feng. 2009. Isolation and partial characterization of novel genes encoding acidic cellulases from metagenomes of buffalo rumens. J. Appl. Microbiol. 107:245-256. [DOI] [PubMed] [Google Scholar]
- 15.Gabor, E. M., E. J. de Vries, and D. B. Janssen. 2003. Efficient recovery of environmental DNA for expression cloning by indirect extraction methods. FEMS Microbiol. Ecol. 44:153-163. [DOI] [PubMed] [Google Scholar]
- 16.Handelsman, J. 2004. Metagenomics: application of genomics to uncultured microorganisms. Microbiol. Mol. Biol. Rev. 68:669-685. (Author's correction, 69:195, 2005.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Handelsman, J., M. R. Rondon, S. F. Brady, J. Clardy, and R. M. Goodman. 1998. Molecular biological access to the chemistry of unknown soil microbes: a new frontier for natural products. Chem. Biol. 5:R245-R249. [DOI] [PubMed] [Google Scholar]
- 18.Heo, S., J. Kwak, H. W. Oh, D. S. Park, K. S. Bae, D. H. Shin, and H. Y. Park. 2006. Characterization of an extracellular xylanase in Paenibacillus sp. HY-8 isolated from an herbivorous longicorn beetle. J. Microbiol. Biotechnol. 16:1753-1759. [Google Scholar]
- 19.Hinman, N. D., D. J. Schell, C. J. Riley, P. W. Bergeron, and P. J. Walter. 1992. Preliminary estimate of the cost of ethanol-production for Ssf technology. Appl. Biochem. Biotechnol 34-35:639-649. [Google Scholar]
- 20.Hinze, B., K. Crailsheim, and R. H. Leuthold. 2002. Polyethism in food processing and social organisation in the nest of Macrotermes bellicosus (Isoptera, Termitidae). Insectes Soc. 49:31-37. [Google Scholar]
- 21.Jiang, Z. Q., W. Deng, Y. P. Zhu, L. T. Li, Y. J. Sheng, and K. Hayashi. 2004. The recombinant xylanase B of Thermotoga maritima is highly xylan specific and produces exclusively xylobiose from xylans, a unique character for industrial applications. J. Mol. Catal. B Enzym. 27:207-213. [Google Scholar]
- 22.Johjima, T., Y. Taprab, N. Noparatnaraporn, T. Kudo, and M. Ohkuma. 2006. Large-scale identification of transcripts expressed in a symbiotic fungus (Termitomyces) during plant biomass degradation. Appl. Microbiol. Biotechnol. 73:195-203. [DOI] [PubMed] [Google Scholar]
- 23.Joshi, M. D., G. Sidhu, I. Pot, G. D. Brayer, S. G. Withers, and L. P. McIntosh. 2000. Hydrogen bonding and catalysis: a novel explanation for how a single amino acid substitution can change the pH optimum of a glycosidase. J. Mol. Biol. 299:255-279. [DOI] [PubMed] [Google Scholar]
- 24.Kim, D. Y., M. K. Han, D. S. Park, J. S. Lee, H. W. Oh, D. H. Shin, T. S. Jeong, S. U. Kim, K. S. Bae, K. H. Son, and H. Y. Park. 2009. Novel GH10 xylanase, with a fibronectin type 3 domain, from Cellulosimicrobium sp. strain HY-13, a bacterium in the gut of Eisenia fetida. Appl. Environ. Microbiol. 75:7275-7279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kvist, T., B. K. Ahring, R. S. Lasken, and P. Westermann. 2007. Specific single-cell isolation and genomic amplification of uncultured microorganisms. Appl. Microbiol. Biotechnol. 74:926-935. [DOI] [PubMed] [Google Scholar]
- 26.Kwon, K. S., J. Lee, H. G. Kang, and Y. C. Hah. 1994. Detection of beta-glucosidase activity in polyacrylamide gels with esculin as substrate. Appl. Environ. Microbiol. 60:4584-4586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lo, N., G. Tokuda, H. Watanabe, H. Rose, M. Slaytor, K. Maekawa, C. Bandi, and H. Noda. 2000. Evidence from multiple gene sequences indicates that termites evolved from wood-feeding cockroaches. Curr. Biol. 10:801-804. [DOI] [PubMed] [Google Scholar]
- 28.Martin, M. M., and J. S. Martin. 1978. Cellulose digestion in the midgut of the fungus-growing termite Macrotermes natalensis: the role of acquired digestive enzymes. Science 199:1453-1455. [DOI] [PubMed] [Google Scholar]
- 29.Matoub, M., and C. Rouland. 1995. Purification and properties of the xylanases from the termite Macrotermes bellicosus and its symbiotic fungus Termitomyces sp. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 112:629-635. [DOI] [PubMed] [Google Scholar]
- 30.Matson, E., E. Ottesen, and J. Leadbetter. 2007. Extracting DNA from the gut microbes of the termite (Zootermopsis nevadensis). J. Vis. Exp. 195:1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller, G. L. 1959. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 31:426-428. [Google Scholar]
- 32.Neufeld, J. D., Y. Chen, M. G. Dumont, and J. C. Murrell. 2008. Marine methylotrophs revealed by stable-isotope probing, multiple displacement amplification and metagenomics. Environ. Microbiol. 10:1526-1535. [DOI] [PubMed] [Google Scholar]
- 33.Ohkuma, M., H. Yuzawa, W. Amornsak, Y. Sornnuwat, Y. Takematsu, A. Yamada, C. Vongkaluang, O. Sarnthoy, N. Kirtibutr, N. Noparatnaraporn, T. Kudo, and T. Inoue. 2004. Molecular phylogeny of Asian termites (Isoptera) of the families Termitidae and Rhinotermitidae based on mitochondrial COII sequences. Mol. Phylogenet. Evol. 31:701-710. [DOI] [PubMed] [Google Scholar]
- 33a.Okada, H., and A. Shinmyo. 1988. Xylanase of Bacillus pumilus. Methods Enzymol. 160:632-637. [Google Scholar]
- 34.Ovreas, L., L. Forney, F. L. Daae, and V. Torsvik. 1997. Distribution of bacterioplankton in meromictic Lake Saelenvannet, as determined by denaturing gradient gel electrophoresis of PCR-amplified gene fragments coding for 16S rRNA. Appl. Environ. Microbiol. 63:3367-3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ragauskas, A. J., C. K. Williams, B. H. Davison, G. Britovsek, J. Cairney, C. A. Eckert, W. J. Frederick, J. P. Hallett, D. J. Leak, C. L. Liotta, J. R. Mielenz, R. Murphy, R. Templer, and T. Tschaplinski. 2006. The path forward for biofuels and biomaterials. Science 311:484-489. [DOI] [PubMed] [Google Scholar]
- 36.Reinholdhurek, B., T. Hurek, M. Claeyssens, and M. Vanmontagu. 1993. Cloning, expression in Escherichia coli, and characterization of cellulolytic enzymes of Azoarcus sp., a root-invading diazotroph. J. Bacteriol. 175:7056-7065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sapag, A., J. Wouters, C. Lambert, P. de Ioannes, J. Eyzaguirre, and E. Depiereux. 2002. The endoxylanases from family 11: computer analysis of protein sequences reveals important structural and phylogenetic relationships. J. Biotechnol. 95:109-131. [DOI] [PubMed] [Google Scholar]
- 38.Schmeisser, C., H. Steele, and W. R. Streit. 2007. Metagenomics, biotechnology with non-culturable microbes. Appl. Microbiol. Biotechnol. 75:955-962. [DOI] [PubMed] [Google Scholar]
- 39.Reference deleted.
- 40.Sogin, M. L., H. G. Morrison, J. A. Huber, D. M. Welch, S. M. Huse, P. R. Neal, J. M. Arrieta, and G. J. Herndl. 2006. Microbial diversity in the deep sea and the underexplored “rare biosphere.” Proc. Natl. Acad. Sci. U. S. A. 103:12115-12120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Subramaniyan, S., and P. Prema. 2002. Biotechnology of microbial xylanases: enzymology, molecular biology, and application. Crit. Rev. Biotechnol. 22:33-64. [DOI] [PubMed] [Google Scholar]
- 42.Teather, R. M., and P. J. Wood. 1982. Use of Congo red polysaccharide interactions in enumeration and characterization of cellulolytic bacteria from the bovine rumen. Appl. Environ. Microbiol. 43:777-780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tokuda, G., N. Lo, H. Watanabe, M. Slaytor, T. Matsumoto, and H. Noda. 1999. Metazoan cellulase genes from termites: intron/exon structures and sites of expression. Biochim. Biophys. Acta 1447:146-159. [DOI] [PubMed] [Google Scholar]
- 45.Tomme, P., R. A. Warren, and N. R. Gilkes. 1995. Cellulose hydrolysis by bacteria and fungi. Adv. Microb. Physiol. 37:1-81. [DOI] [PubMed] [Google Scholar]
- 46.Voget, S., H. L. Steele, and W. R. Streit. 2006. Characterization of a metagenome-derived halotolerant cellulase. J. Biotechnol. 126:26-36. [DOI] [PubMed] [Google Scholar]
- 47.Wakarchuk, W. W., R. L. Campbell, W. L. Sung, J. Davoodi, and M. Yaguchi. 1994. Mutational and crystallographic analyses of the active-site residues of the Bacillus circulans xylanase. Protein Sci. 3:467-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Walfridsson, M., X. M. Bao, M. Anderlund, G. Lilius, L. Bulow, and B. Hahn-Hagerdal. 1996. Ethanolic fermentation of xylose with Saccharomyces cerevisiae harboring the Thermus thermophilus xylA gene, which expresses an active xylose (glucose) isomerase. Appl. Environ. Microbiol. 62:4648-4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Warnecke, F., P. Luginbuhl, N. Ivanova, M. Ghassemian, T. H. Richardson, J. T. Stege, M. Cayouette, A. C. McHardy, G. Djordjevic, N. Aboushadi, R. Sorek, S. G. Tringe, M. Podar, H. G. Martin, V. Kunin, D. Dalevi, J. Madejska, E. Kirton, D. Platt, E. Szeto, A. Salamov, K. Barry, N. Mikhailova, N. C. Kyrpides, E. G. Matson, E. A. Ottesen, X. N. Zhang, M. Hernandez, C. Murillo, L. G. Acosta, I. Rigoutsos, G. Tamayo, B. D. Green, C. Chang, E. M. Rubin, E. J. Mathur, D. E. Robertson, P. Hugenholtz, and J. R. Leadbetter. 2007. Metagenomic and functional analysis of hindgut microbiota of a wood-feeding higher termite. Nature 450:560-565. [DOI] [PubMed] [Google Scholar]
- 50.Watanabe, H., H. Noda, G. Tokuda, and N. Lo. 1998. A cellulase gene of termite origin. Nature 394:330-331. [DOI] [PubMed] [Google Scholar]
- 51.Wegley, L., R. Edwards, B. Rodriguez-Brito, H. Liu, and F. Rohwer. 2007. Metagenomic analysis of the microbial community associated with the coral Porites astreoides. Environ. Microbiol. 9:2707-2719. [DOI] [PubMed] [Google Scholar]
- 52.Wood, T. G., and R. J. Thomas. 1989. The mutualistic association between Macrotermitinae and Termitomyces, p. 69-92. In N. Wilding, N. M. Collins, P. M. Hammond, and J. F. Webber (ed.), Insect-fungus interaction. Academic Press, London, England.
- 53.Yamada, A., T. Inoue, D. Wiwatwitaya, M. Ohkuma, T. Kudo, T. Abe, and A. Sugimoto. 2005. Carbon mineralization by termites in tropical forests, with emphasis on fungus combs. Ecol. Res. 20:453-460. [Google Scholar]
- 54.Yu, J. H., Y. S. Park, D. Y. Yum, J. M. Kim, I. S. Kong, and D. H. Bai. 1993. Nucleotide sequence and analysis of a xylanase gene (xynS) from alkali-tolerant Bacillus sp. YA-14 and comparison with other xylanases. J. Microbiol. Biotechnol. 3:139-145. [Google Scholar]
- 55.Zhang, K., A. C. Martiny, N. B. Reppas, K. W. Barry, J. Malek, S. W. Chisholm, and G. M. Church. 2006. Sequencing genomes from single cells by polymerase cloning. Nat. Biotechnol. 24:680-686. [DOI] [PubMed] [Google Scholar]
- 56.Zhang, Z. Q., et al. 2005. Thin layer chromatography analysis of several uronates and their oligosaccharides. Chin. J. Analyt. Chem. 33:1750-1752. [Google Scholar]
- 57.Zhou, J. Z., M. A. Bruns, and J. M. Tiedje. 1996. DNA recovery from soils of diverse composition. Appl. Environ. Microbiol. 62:316-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.