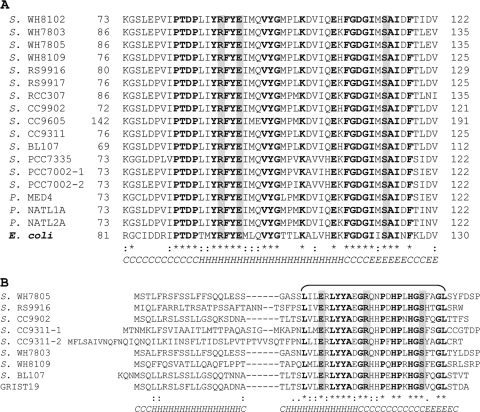
FIG. 9.
(A and B) Amino acid alignments for catalytic site regions of cyanases for 17 CynS sequences (A) and 9 full-length CynH sequences (B) found in marine Synechococcus (S.) and in a single metagenomic clone GRIST19. Identical residues are presented in boldface and labeled “*”, conserved residue replacements are labeled “:”, and functionally similar residues are labeled “.”. The proposed active-site residues Arg (R), Glu (E), and Ser (S) are shown in boldface against a gray background. The bottom line presents the consensus secondary structure predicted by the Jpred prediction server that identifies randomly coiled region (C), α-helix (H), and β-sheet (E) motifs. In panel B, the aligned sequences are cyanate hydratase (ZP_01124909) in Synechococcus sp. WH7805, RS9916_37357 (ZP_01471502) in Synechococcus sp. RS9916, Syncc9902_2288 (YP_378289) in Synechococcus sp. CC9902, sync_2840 and sync_2903 (YP_732028 and YP_732090) in Synechococcus sp. CC9311, SynWH7803_2496 (YP_001226219) in Synechococcus sp. WH7803, SH8109_0550 (ZP_05789530) in Synechococcus sp. WH8109, non-annotated (reverse strand 1870903-1871064) in Synechococcus sp. BL107 (NZ_AATZ00000000), and metagenomic clone GRIST19 (EU795157).