Abstract
In Escherichia coli, the aerotaxis receptor Aer is an atypical receptor because it senses intracellular redox potential. The Aer sensor is a cytoplasmic, N-terminal PAS domain that is tethered to the membrane by a 47-residue F1 linker. Here we investigated the function, topology, and orientation of F1 by employing random mutagenesis, cysteine scanning, and disulfide cross-linking. No native residue was obligatory for function, most deleterious substitutions had radically different side chain properties, and all F1 mutants but one were functionally rescued by the chemoreceptor Tar. Cross-linking studies were consistent with the predicted α-helical structure in the N-terminal F1 region and demonstrated trigonal interactions among the F1 linkers from three Aer monomers, presumably within trimer-of-dimer units, as well as binary interactions between subunits. Using heterodimer analyses, we also demonstrated the importance of arginine residues near the membrane interface, which may properly anchor the Aer protein in the membrane. By incorporating these data into a homology model of Aer, we developed a model for the orientation of the Aer F1 and PAS regions in an Aer lattice that is compatible with the known dimensions of the chemoreceptor lattice. We propose that the F1 region facilitates the orientation of PAS and HAMP domains during folding and thereby promotes the stability of the PAS and HAMP domains in Aer.
Escherichia coli bacteria navigate to microenvironments where the oxygen concentration, energy sources, and redox potential are optimal for growth. This process is in part orchestrated by the aerotaxis receptor Aer, which measures redox potential and infers energy levels via a flavin adenine dinucleotide (FAD) cofactor bound to a cytoplasmic PAS domain (8, 9, 15, 29, 30, 33, 34). A decrease in oxygen lowers the redox potential and reduces PAS-FAD, initiating conformational changes that propagate through the Aer HAMP domain to the kinase control module (33). The activated module increases the autophosphorylation rate of the CheA histidine kinase and, in turn, the phosphorylation levels of the response regulator CheY, which binds to the flagellar motor, causing clockwise rotation and tumbling (see reference 18 for a review).
The Aer receptor has two cytoplasmic segments that are anchored in the membrane by a hairpin loop. The N-terminal PAS domain is tethered to the membrane anchor by a 47-amino-acid region known as F1. This region has an unknown structure with low sequence conservation, but it is known to influence protein stability (11) and is indispensable for the function of Aer (39). Heterodimers composed of one full-length Aer monomer and one truncated monomer can function, providing that the truncated monomer contains native Aer residues 120 to 506 (39). Thus, the PAS domain (residues 1 to 119) from one of the monomers is dispensable, but both F1 segments (residues 120 to 166 [8]) are essential.
Aer and chemotaxis receptors in E. coli have similar HAMP domains and kinase control modules, but the sensing region in the chemotaxis proteins (Tsr, Tar, Trg, Tap) is in the periplasm, whereas the sensor is in the cytoplasm in Aer. Despite these differences, chemoreceptors, as well as Aer, form homodimers that assemble into mixed squads of trimers of dimers with little or no preference for self (2, 16). Each trimer-of-dimer squad forms a signaling team with CheW and CheA proteins, and the teams are regularly positioned as vertices in a hexagonal lattice that has a fixed center-to-center spacing of 12 nm (10). These units constitute a larger structural lattice containing thousands of receptors at the cell poles, giving a honeycomb appearance when viewed by electron cryotomography (10). Although the orientation between Aer dimers in a trimer of dimers is not known, the relative positioning is probably similar in both homogeneous and mixed trimers of dimers because the kinase control module limits rotational freedom at the membrane (4).
Recently, we reported that the β scaffolding of the Aer PAS contacts the AS-2 helix of the HAMP domain (12). In principle, it is possible that the F1 regions promote stability by correctly orienting the PAS and HAMP domains and/or assisting the proper vertical registry. In this study, we investigated the F1 region by employing random mutagenesis, cysteine scanning, and disulfide cross-linking. The cross-linking data were then used to predict the orientation of F1 and PAS in an Aer lattice.
MATERIALS AND METHODS
Bacterial strains and plasmids.
E. coli strains BT3388 (Δaer::erm Δtsr-7021 Δtar-tap-5201 trg::Tn10) (43), BT3312 (Δaer-1 Δtsr-7021) (30), and BT3400 (Δaer-1 Δtsr-7021 recA::cat) (39) were used in this study. These strains were derived from E. coli RP437, which is wild type for chemotaxis (27).
The plasmids used in this study included pTrc99A (Pharmacia, Piscataway, NJ), an isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible ptrc expression vector; pGH1 (29), a pTrc99A derivative expressing wild-type Aer; pMB1 (26), a pGH1 derivative that expresses cysteineless (C-less) Aer (Aer-C193S/C203S/C253A); pKW1 (41), a pGH1 derivative containing silent mutations that introduce restriction sites in aer for NheI (codon 14), BstBI (codon 204), and SacI (codon 281); pProEX HTa (Gibco BRL, Life Technologies, Gaithersburg, MD), an IPTG-inducible ptrc expression vector; pAVR1 (21), a pProEX-based vector expressing His6-tagged Aer2-166; pKW94 (39), a pACYC184-derived plasmid expressing AerQ248R under the control of the IPTG-inducible ptrc promoter; pDS7 (39), which expresses wild-type Aer from pACYC184 using a tightly regulated sodium salicylate-inducible promoter (pnahG); and pLC113 (a gift from J. S. Parkinson) (1), a pACYC184-based plasmid that expresses wild-type Tar and carries a sodium salicylate-inducible promoter. pACYC184 (13) contains a p15A origin of replication, allowing coexpression of genes with pTrc99a- or pProEx-derived plasmids.
Mutagenesis.
Site-directed cysteine mutagenesis of the region encoding Aer residues 122 to 166 was performed according to the instructions of the QuikChange site-directed mutagenesis kit (Agilent Technologies, Santa Clara, CA) using pMB1 as the template. To obtain single amino acid changes in the F1 region, random PCR mutagenesis was performed using pGH1 and primers NheIF and BstBIR (41). Both Taq (Fisher Scientific, Pittsburgh, PA) and Mutazyme II (Agilent Technologies, Santa Clara, CA) DNA polymerases were used. Reaction mixtures containing Taq DNA polymerase were prepared under conditions of reduced fidelity (22) as previously described (41). The Taq-generated DNA fragments were then pooled and purified using a QIAquick PCR purification column (Qiagen Inc., Valencia, CA) and subjected to 30 cycles of normal PCR amplification (95°C for 30 s, 59°C for 30 s, and 72°C for 30 s). Random PCR mutagenesis using Mutazyme II DNA polymerase was performed according to the instructions of the Genemorph II random mutagenesis kit (Agilent Technologies). PCR products obtained by both methods were purified, digested with NheI and BstBI, and then gel purified using a QIAquick gel extraction kit (Qiagen). The digested fragments were subsequently cloned into pKW1 with the corresponding NheI/BstBI fragment removed and then introduced into BT3388 by electroporation. Aer expression was confirmed by Western blot analysis using anti-Aer2-166 antiserum (30), and mutations were confirmed by sequencing the entire aer gene.
Construction of truncation mutants.
N-terminal Aer truncations Δ1-126, Δ1-134, and Δ1-153 were constructed by PCR using pGH1 as the template. Sense primers complementary to the NcoI site at pTrc99A nucleotide 265 were paired with an antisense primer containing a SalI site. The sense primers incorporated a start codon followed by the codon for residue 127, 135, or 154, while the AerSalI antisense primer contained residue 506 preceded by the normal stop codon of Aer. The PCR products were digested with NcoI and SalI and then cloned into pTrc99A. The N-terminal truncations were verified by DNA sequencing, and protein expression was confirmed by Western blot assay using anti-Tsr antiserum (supplied by Claudia Studdert, Universidad Nacional de Mar del Plata, Argentina). DNA encoding Aer165-506 (39) was also cloned into pProEx, which introduces a 24-amino-acid, unstructured, soluble leader into the N terminus.
Mutant characterization.
Individual BT3312 Aer single-cysteine mutants were analyzed for aerotaxis at 30°C in minimal semisolid agar containing 30 mM succinate and 50 μg ml−1 ampicillin (33). BT3388 Aer F1 mutants (derived by random mutagenesis) were screened for aerotaxis defects at 30°C in Tryptone semisolid agar miniswarm plates (11) containing 0.35% Bacto agar (Difco Laboratories, Sparks, MD), 50 μg ml−1 ampicillin, and 20 μM IPTG. Aerotaxis-defective colonies were confirmed in Tryptone semisolid agar plates (33) containing 50 μg ml−1 ampicillin and 20 to 1,000 μM IPTG. Plasmids from aerotaxis-defective mutants were then expressed in BT3312 and assessed in minimal succinate semisolid agar with 50 μg ml−1 ampicillin and 0 to 1,000 μM IPTG. Mutants that remained nonaerotactic in succinate semisolid agar with up to 1,000 μM induction were tested in BT3400 for dominant and recessive effects on wild-type Aer as previously described (42). The nonaerotactic BT3388 Aer F1 mutants (derived by random mutagenesis) were also assessed in a gas perfusion chamber for their response to oxygen after induction with 200 or 1,000 μM IPTG as described previously (29).
Steady-state mutant protein levels were determined by Western blot assay after growing BT3388 Aer F1 mutants in Luria-Bertani “lysogeny broth” (LB) (7) plates with 0.5 μg ml−1 thiamine (LB thia) medium to mid-log phase at 30°C and inducing with 50 μM IPTG for 3 h. The band intensity of each mutant protein was compared to that of wild-type Aer as expressed from pKW1. Protein degradation assays were performed by inducing cells grown to mid-log phase in LB thia with 50 μM IPTG for 3 h and then blocking protein synthesis with 500 μg ml−1 chloramphenicol. Samples were collected at different time points over 4 h and analyzed for full-length Aer expression by Western blot assay. Densitometry measurements were done using a UVP Biospectrum 500 Multispectrum Imaging System (Upland, CA).
Heterodimer complementation assays.
BT3400 cells were cotransformed with plasmids expressing AerQ248R (pKW94, pACYC184 derived [39]) and one of the N-terminally truncated Aer receptors (pTrc99A or pProEX derived). Cotransformants were selected on LB thia and 100 μg ml−1 ampicillin as previously described (39). BT3400 cells were also transformed with the relevant plasmids individually to create appropriate homodimer controls. Heterodimer aerotaxis assays were performed in minimal semisolid agar containing 30 mM succinate, 7.5 μg ml−1 tetracycline, and 50 μg ml−1 ampicillin (39). Both plasmids were IPTG inducible, and gene expression levels were varied by adding IPTG to the semisolid agar in a series of titrations (0, 20, 50, 100, 200, 400, 600, and 1,000 μM). Appropriate homodimer controls were carried out simultaneously with every heterodimer experiment.
Dominance testing.
To test for dominant behavior, mutant Aer constructs were cotransformed with pDS7, which expresses wild-type Aer from pACYC184 using a tightly regulated sodium salicylate-inducible promoter (nahG) (39). Dominance tests were performed the same way as the heterodimer assays outlined above, except that 0.5 or 1 μM sodium salicylate and IPTG concentrations between 0 and 0.6 mM were included in the swarm plates in a series of titrations. Induction levels producing approximately 1:1 expression ratios were determined by Western blot assay.
In vivo cross-linking.
Disulfide cross-linking was performed at 25°C by exposing BT3312 cells to 300 μM copper phenanthroline as previously described (5, 33, 40). Cross-linking was also performed at 25°C by treating BT3312 cells with 6 μg ml−1 of bis-maleimidoethane (BMOE; Pierce, Rockford, IL) and then incubating them for 0, 2, 5, 10, or 15 min. Reactions were quenched with stop solution containing 80 mM β-mercaptoethanol, and cells were lysed by boiling for 4 min. Aer monomer and dimer bands were visualized after sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting using an Alpha Innotech digital imaging system.
Similarly, induced intact cells were treated with Tris(2-maleimidoethyl)amine (TMEA; Pierce Biotechnologies, Rockford, IL). Cells were harvested by centrifugation, resuspended in KEP buffer (2), and then treated with 50 μM TMEA for 15 min at 30°C. Reactions were quenched with 10 mM N-ethylmaleimide (NEM). Cells were pelleted by centrifugation and lysed by boiling in stop solution containing 10 mM NEM for 4 min (32). Monomer, dimer, and trimer bands were separated by SDS-PAGE.
Detergent extraction of Aer fragments from the membrane.
BT3388 membrane fractions expressing various Aer fragments were isolated as previously described (19). Briefly, membrane fractions were treated with 2% mild detergent Triton X-100, incubated on ice for 30 min, and then centrifuged at 485,000 × g for 30 min to remove nonaggregated protein from the membrane. This process was repeated, and aliquots from each fraction were Western blotted. BT3388 cells expressing pGH1 or pAVR1 were used as positive and negative controls, respectively.
Protein prediction algorithms.
A consensus for the secondary structure of the Aer-F1 region (residues 120 to 166) was generated using the SYMPRED program (http://www.ibi.vu.nl/programs/sympredwww/; Centre for Integrative Bioinformatics VU, University of Amsterdam). The program employed three rounds of PSI-BLAST to produce 72 alignments that were used as the input for the prediction programs. A consensus was generated with dynamic programming and no weighting using a combination of the following programs integrated into SYMPRED: PHDpsi (31), PROFsec (B. Rost, unpublished data), SSPro2.01 (28), YASPIN (25), Jnet (14), and PSIpred (21).
Sequence alignment of the E. coli Aer PAS/N-terminal F1 region and MmoS-PAS-A from Methylococcus capsulatus (Bath) (36) was done with ClustalW (23).
RESULTS
Influence of F1 substitutions on Aer function.
To assess the functional requirement of each native amino acid in the F1 region, we made single cysteine substitutions in residues 120 to 166 by mutating codons in C-less Aer expression vector pMB1. We chose cysteine because it has an average size, it can be accommodated in both hydrophobic and hydrophilic environments, and it can be targeted in related biochemical experiments to determine solvent accessibility or proximity between domains (6, 35, 40). Mutant proteins with single cysteine substitutions were expressed in E. coli BT3312 (aer tsr), which lacks both aerotaxis receptors, Aer and Tsr (29). In BT3312, all Cys-substituted Aer proteins supported aerotaxis and exhibited normal morphologies in succinate semisolid agar. One mutant protein, Aer-L146C, generated a larger-than-normal “superswarming” colony with a swarm rate of 140% of the wild-type rate. Since all of the engineered Aer proteins mediated aerotaxis, we conclude that the cysteine substitutions did not significantly distort the native fold of Aer. Moreover, no single native residue in this region is obligatory for Aer signaling.
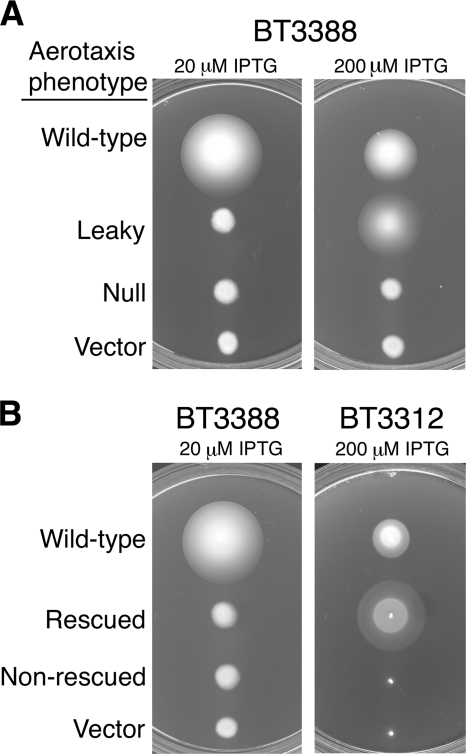
In a previous mutagenesis study, Burón-Barral and coworkers identified seven substitutions at six sites within the F1 region that produced unstable Aer proteins with a defective aerotaxis phenotype (11). In the present study, we employed low-fidelity PCR to randomly introduce single amino acid substitutions into the F1 region and further explored the role of F1 in the Aer protein. To reduce mutational bias, we used both Taq and Mutazyme II DNA polymerases (37). We expressed the mutant proteins in receptorless BT3388 (tar tsr trg tap aer) and screened on Tryptone semisolid agar for nonaerotactic colonies having low expansion rates and lacking an outer aerotaxis ring. Those mutants that produced small amounts of Aer product (estimated by Western blot assays after induction with 1 mM IPTG) were discarded, leaving 10 abnormal isolates with substitutions at six previously unidentified sites. The new substitutions included A124P, A135P, G145V, G154D, R161P, R162P, and R166P, while substitutions previously reported included I123N, I142T, and L161H (11). To test for leaky Aer mutants that have partial function, the expression of the 10 mutant receptors was induced incrementally with IPTG (20 to 1,000 μM) (Fig. 1A). At higher induction levels, five of the mutants (A135P, I142T, G154D, L161H, and R162P) had partial Aer function on Tryptone semisolid agar (leaky, Fig. 1A), but the remaining mutants displayed nonaerotactic phenotypes at all induction levels (null, Fig. 1A) and failed to tumble like wild-type Aer when nitrogen replaced oxygen in a gas perfusion assay (data not shown).
FIG. 1.
Aerotaxis phenotypes of Aer cysteine mutants. (A) Mutant Aer proteins were expressed in receptorless strain BT3388 (aer tsr tar tap trg), and colonies were inoculated onto a series of Tryptone semisolid agar plates containing 50 μg ml−1 ampicillin and the IPTG concentration specified. Plates were incubated for 16 h. The examples shown are for plasmids pKW1 (wild-type Aer), pKW1-A135P (leaky), pKW1-A124P (null), and pTrc99A (vector). (B) Plasmids isolated from aerotaxis-defective colonies in panel A were introduced into BT3312 (aer tsr) to test for functional rescue by Tar. Colonies were inoculated onto succinate semisolid agar plates containing 50 μg ml−1 ampicillin and the IPTG concentration shown, and plates were incubated for 18 to 19 h. The examples shown are for plasmids pKW1 (wild-type Aer), pKW1-A124P (rescued), pKW1-I123N (nonrescued), and pTrc99A (vector).
Some defective Aer proteins can be functionally rescued by the presence of the high-abundance chemoreceptor Tar, indicating that these aberrant Aer receptors retain some input/output control (11, 16). The mechanism of rescue is unclear but may be a combination of Tar correction of tumbling bias and the ability of Tar to exert compensatory effects on the structure of Aer through direct interactions within a mixed trimer of dimers (16). When expressed in the presence of the Tar high-abundance chemoreceptor (in BT3312 [aer tsr]), four of the five remaining F1 mutants were functionally rescued (Fig. 1B). The only Aer mutant that was not functionally rescued by Tar was Aer-I123N; this mutant was also phenotypically recessive, since it did not alter wild-type-mediated aerotaxis when coexpressed (at a 1:1 ratio) with wild-type Aer.
Proteolysis of the F1 mutant proteins.
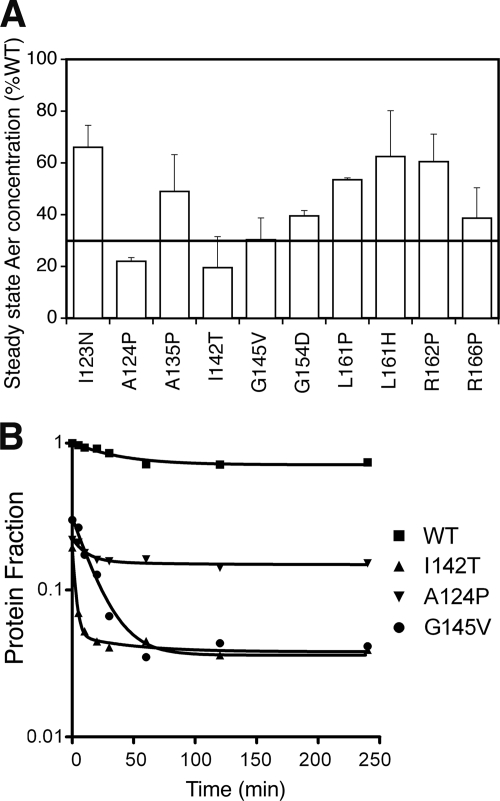
A previous study found that Aer proteins with F1 substitutions were rapidly proteolyzed (11), and we examined the influence of the present F1 substitutions on protein stability by determining the steady-state accumulation level of each mutant protein after induction with 50 μM IPTG. Steady-state levels ranged from 20% to 67% of that of wild-type Aer (pKW1) (Fig. 2A), and those with levels below 30% were assayed for proteolysis. Compared to wild-type Aer, which was stable and had a low initial degradation rate (0.007/min) and little or no subsequent degradation, the F1 mutant proteins had significant initial rates of degradation (Fig. 2B). The three mutants with rapid degradation included Aer-A124P (0.0315/min), Aer-G145V (0.0533/min), and Aer-I142T (0.1733/min). The initial rapid decay of the mutant proteins was followed by longer periods of slow decay (Fig. 2B) and likely reflects two protein populations: partially folded proteins that had not completed the maturation process and mature proteins that were more stable (11).
FIG. 2.
Steady-state accumulation and degradation profiles of mutant Aer proteins. (A) Steady-state concentrations of mutant Aer proteins relative to wild-type (WT) Aer when induced with 50 μM IPTG in receptorless strain BT3388. Mutant proteins that were present at less than 30% of normal (line) were chosen for degradation assays. (B) Degradation profiles of mutant Aer proteins. Samples were collected at selected time points after inhibition of new protein synthesis with chloramphenicol, and Aer levels were quantified by Western blot assay. The degradation curves represent best-fit values for a two-phase exponential decay, and the values shown are fractional concentrations relative to wild-type Aer.
Cross-linking to assess F1 proximity and structure.
The finding that the F1 substitutions alter stability more than function suggests that the F1 region is not intimately involved in Aer signal transduction and probably lends structural stability to the PAS and/or HAMP domains. In principle, this stability could be provided directly through F1-F1, F1-PAS, or F1-HAMP contacts or conferred indirectly by influencing PAS-HAMP positioning or membrane insertion.
To assess whether the cognate F1 regions form direct contacts, we employed in vivo experiments using the Cys-substituted Aer receptors. For each of the engineered Aer proteins, we determined the extent of cysteine cross-linking in whole cells 10 min after adding the oxidant copper phenanthroline (CuPhe) (35). Cross-linked Aer dimers were separated from monomers by SDS-PAGE under nonreducing conditions and visualized by Western blot assay. Only six cysteine substitutions cross-linked to form Aer dimers (Fig. 3A). One contact site was located at the N terminus (E121C), three sites were in the center (A135C, G136C, and R137C), and two were at the C terminus (A165C and R166C) of the F1 region. The sparse cross-linking indicates that the F1 regions are not proximal and do not form a continuous interacting face like the AS-2 helices in the Aer HAMP domain (40).
FIG. 3.
Percentage of dimer formation for each Aer cysteine substation in the F1 region. (A) Dimers formed by disulfide bonds after oxidation of whole cells with CuPhe for 10 min at 25°C. (B) Dimers formed by incubating whole cells with the bifunctional, sulfhydryl-reactive probe BMOE. A consensus secondary structure (SYMPRED program; http://www.ibi.vu.nl/programs/sympredwww/; Center for Integrative Bioinformatics VU, University of Amsterdam) is shown above the graph, where rectangles indicate helices, lines indicate coils (loops), and arrows indicate β strands. The open circles under the secondary-structure prediction indicate the locations of missense mutations that produced mutant proteins (from Fig. 2A). An N-terminal helical model is shown to highlight the predicted locations of three nonreactive residues in the helix (V126, L129, and L133; sticks facing downward). The helix is a homology model based on the region in MmoS from M. capsulatus with sequence similarity (see Fig. S1 in the supplemental material).
Cross-linking with BMOE.
To extend the “reach” of the individual Cys residues and further explore the structure of F1, we employed the molecular ruler BMOE, which is a homobifunctional thiol-reactive probe with a dynamic simulated average length of 8.18 ± 0.75 Å (17). This probe can readily enter intact cells and cross-link cysteine residues that lie within its dynamic range (6.27 to 10.52 Å [17]) (3, 35). In these studies, we compared the extent of cross-linking after 5 min because cross-linking levels reached a plateau before 10 min for some of the cysteine residues (data not shown). Using the BMOE probe, 18 of the 47 Cys replacements cross-linked (Fig. 3B). Compared with a consensus secondary structure generated by SYMPRED, several notable features emerged (Fig. 3B). The N-terminal F1 region was predicted to be an α helix, and this structure is consistent with the cross-linking pattern of residues 126 to 133. When mapped onto a helical model, the unreactive residues V126C, L129C, and L133C lay on the same face, projecting in the direction opposite to that of the reactive residues (Fig. 3B). Residue L133 was followed by six consecutive cross-linked residues (residues 134 to 139), indicating that the next segment is dynamic and probably a loop. Notably, the consensus secondary structure predicted a slightly smaller loop extending from residue 136 to residue 140 (Fig. 3A and B). In the 11-amino-acid segment following residue 139, only one substitution (L146C) cross-linked (Fig. 3B). This position was predicted to lie at the beginning of a β strand, but in the absence of a cross-linking pattern, no inferences could be drawn. Moreover, Aer-L146C mediated a superswarming phenotype (see above), so the precise orientation of the native residue might be different from the Cys substitution. Finally, three more substitutions cross-linked in consecutive positions from 150 to 152, supporting the consensus prediction that these residues form a loop (150 to 153) (Fig. 3B). Unexpectedly, residues A165C and R166C did not cross-link with BMOE, even though the residues form cognate disulfide bonds when oxidized with CuPhe (see above). The reason for this is unclear, but these residues may be closer together than the dynamic range of the probe (17), or the environment at the membrane/cytosol interface may not be favorable for cross-linking with this probe.
Placement of the F1 region within a trimer of dimers.
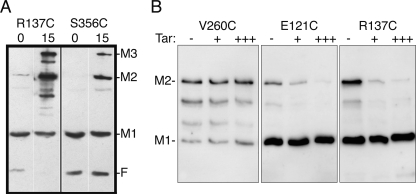
In wild-type E. coli, Aer forms mixed trimers of dimers with the four chemoreceptors, but in the absence of chemoreceptors in a receptorless strain, plasmid-borne Aer forms homogeneous trimers of dimers (4, 16). Since the Aer signaling region governs dimer-dimer orientation (4), the relative positioning of Aer is likely similar in both homogeneous and mixed trimers of dimers. To probe the positioning of the F1 region within a trimer of Aer dimers, we used the trifunctional thio-reactive probe TMEA, which is capable of entering cells and covalently binding to three sulfhydryls that lie within 10.3 Å of one another (Pierce) (32). For this test, we used Aer-R137C, which exhibited the highest rate of disulfide formation with CuPhe and probably lies within a loop (see above). Receptorless BT3388 cells expressing Aer-R137C were incubated with TMEA for 15 min, the reaction was stopped with NEM, and denatured products were separated by SDS-PAGE. Western blot assays showed a band of Aer protein with the mobility of a trimer (Fig. 4A), indicating that the probe cross-linked three monomers. This suggested that the loop regions of three F1 monomers are in close proximity and perhaps face a pocket circumscribed by the trimer of dimers. In this case, disulfide cross-links between F1 regions might occur between dimers, as opposed to (or in a addition to) within dimers.
FIG. 4.
Spatial proximity among three F1 monomers from different dimers. (A) The trifunctional sulfhydryl-reactive cross-linker TMEA (with three 10.3-Å spacers) cross-linked three AerR137C monomers. Substitution R137C is in a loop region of F1 (Fig. 3). S356C is known to cross-link with TMEA (16, 32) and was used as a positive control. Combinations of cross-linked monomers and the fragment are evident in lane 2 but not in lane 4, indicating that the C-terminal half of the fragment did not cross-link (S356C is C terminal to the cleavage site). Anti-Aer2-166 antibodies bind to the N-terminal, but not the C-terminal, fragment. Plasmid-bearing BT3312 cells were incubated with 50 μM TMEA for 15 min at 30°C (lanes 2 and 4) before quenching with 10 mM NEM. Untreated cells were quenched immediately as zero time controls (lanes 1 and 3). F is a proteolytic fragment of Aer formed in vivo (26). M1, M2, and M3 indicate the number of monomers cross-linked. (B) Influence of Tar titrations on the extent of disulfide formation for target Cys substitutions in Aer. Tar decreased the extent of disulfide formation between Cys substitutions in the F1 region (E121C, A135C [not shown], and R137C), but not in the proximal signaling domain (V260C), where contacts are known to be intradimeric. Lanes: −, cells without Tar; +, cells containing tar-inducible plasmid pLC113 (1) without induction; +++, induced with 1.2 μM sodium salicylate.
To differentiate intra- from interdimeric disulfides, we decreased the probability of collisions between Aer dimers by titrating cells with wild-type Tar. Tar joins Aer in mixed trimers of dimers and therefore should decrease the extent of cross-linking between, but not within, Aer dimers. Representative Aer-Cys mutations E121C, A135C, and R137C were introduced into chemoreceptorless BT3388 (43) and expressed alone or with a compatible plasmid harboring a salicylate-inducible tar gene (1). F1 disulfide formation was initiated by exposing cells to CuPhe for 10 min. Under these conditions, the proportion of cross-linked Aer diminished with increasing Tar expression, indicating that collisions between neighboring F1 regions had decreased (Fig. 4B). In contrast, the extent of cross-linking for a known intradimeric cross-linker (V260C, in the proximal signaling region) was not altered by increased Tar expression (Fig. 4B).
Essential arginines in the F1 region.
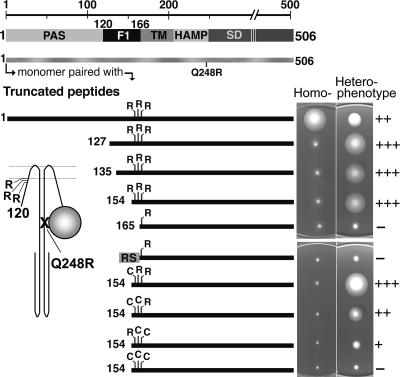
Aer heterodimers can mediate aerotaxis when one monomer is full length and the other monomer lacks the PAS domain, but heterodimers are nonfunctional when the truncated monomer lacks both the PAS and F1 regions (39). These findings indicate that part of the F1 domain is required for aerotaxis. To investigate which segment of the F1 domain is needed for aerotaxis, we measured aerotaxis using Aer N-terminal truncations with nested deletions in the F1 domain (Fig. 5). The heterodimers used in these studies were synthesized in BT3400 (aer tsr recA) from compatible plasmids that expressed (i) a full-length Aer monomer with a Q248R substitution in the HAMP domain and (ii) a truncated Aer peptide with the wild-type HAMP sequence and a segment of F1 (Fig. 5). Homodimers of Aer fragments did not support aerotaxis (Fig. 5) because both PAS domains were deleted; homodimers of full-length AerQ248R peptides did not support aerotaxis because the HAMP domain was defective in both monomers (data not shown) (39). However, the heterodimers supported aerotaxis providing that a segment of the F1 domain was present in the Aer fragment (Fig. 5, upper panels). Thus, all heterodimers supported aerotaxis, with the exception of Aer165-506, suggesting that F1 residues within the 154-to-164 region were essential for aerotaxis (Fig. 5). An alternative possibility is that the segment is necessary to assist or stabilize the membrane anchor, since residue 165 resides at the membrane-cytosol boundary and may need a cytosolic leader to be properly tethered. In this case, any soluble sequence could potentially substitute in this role. To test this possibility, we replaced the PAS and F1 residues with a soluble unstructured 24-amino-acid random sequence (RS, Fig. 5) ligated to residue 165 at the transmembrane junction (Fig. 5, lower panels). When incrementally expressed in BT3400 (20 to 1,000 μM IPTG), this heterodimer did not support aerotaxis in succinate semisolid agar (Fig. 5).
FIG. 5.
Summary of the organization of the Aer domain, the heterodimer constructs, and the phenotypes of these constructs in succinate semisolid agar. The cartoon represents an Aer heterodimer with one peptide N terminally truncated to the F1 region and the other monomer as a defective full-length protein with a Q248R substitution. The locations of the three arginines that may contribute to membrane anchoring are also represented. N-terminal deletions were tested for the ability to rescue the full-length mutant Aer protein containing the Q248R replacement. Their functionality, as well as the influence of the arginines and that of a random 25-residue sequence (RS), is summarized by the phenotypes they produce on succinate semisolid agar (right). Proteins were expressed from compatible plasmids in BT3400 (aer tsr recA). Transformants were inoculated onto succinate semisolid agar plates containing 7.5 μg ml−1 tetracycline and 50 μg ml−1 ampicillin. Left panel, phenotypes corresponding to homodimers identified in the far left column; right panel, phenotypes corresponding to heterodimers composed of full-length AerQ248R and the peptide identified in the left panel. Phenotypes were graded on a scale ranging from − for no function to +++ for maximum colony size. Both plasmids were IPTG inducible, and as shown, induction was 0 μM for the homodimers and 1,000 μM for the heterodimers. Homodimers of AerQ248R or of the N-terminal truncations were nonaerotactic at all induction levels up to 1,000 μM. Induction was unnecessary for all but two of the heterodimers labeled + and ++ in the bottom right panel; colonies formed by bacteria producing full-length heterodimers (top right panel) were larger at lower induction (not shown), and heterodimers labeled − were nonfunctional at all IPTG levels.
We considered the possibility that specific Aer residues within the 154-to-164 segment have a role in anchoring the receptor in the membrane. Residues typically important for insertion and membrane anchoring are positively charged cytosolic residues (24, 38); this F1 segment has three arginine residues at positions 162, 164, and 166, and two of these arginines are missing from Aer165-506 (Fig. 5). We tested the influence of the arginines on function by constructing mono-, di-, and tricysteine replacements of arginines at residues 162, 164, and 166 in both Aer154-506 and full-length Aer (pMB1) (Fig. 5). The constructs were tested as homodimers and heterodimers and compared with full-length Aer homodimers containing identical cysteine replacements. Truncated proteins with one or two arginine residues mediated aerotaxis in heterodimers, but those lacking all three arginines did not (Fig. 5). Notably, the three arginine residues did not appear to be equivalent in supporting aerotaxis (R166 > R164 > R162) (Fig. 5, bottom plates). This gradation of responses was also true for full-length homodimeric Aer, where one arginine was required and the hierarchy was also R166 > R164 > R162 (data not shown). Perhaps residues R164 and R162 did not anchor the Aer protein in the membrane at the same position as R166.
Considering that R162, R164, and R166 are likely to be important in anchoring the Aer protein in the membrane, the absence of all of these arginine residues could affect the topology of Aer, cause misfolding, and result in the formation of inclusion bodies in the cells. We previously observed that misfolded Aer peptides are not readily solubilized by mild detergents and likely form inclusion bodies (19). We tested this possibility in the present study and found that all Aer peptide fragments associated with the membrane and were readily solubilized in 1% Triton X-100 (data not shown). However, we did not investigate the location of the N-terminal region, which may have been exported into the periplasm, even though there was no evidence of gross misfolding (24, 38).
DISCUSSION
All Aer proteins with single Cys substitutions in F1 were functional, indicating that no single residue in this region is indispensable for function. Moreover, all aerotaxis-negative mutants isolated after random mutagenesis had substitutions in the Aer protein known to disrupt protein structure: five mutations introduced a proline, two replaced native glycines, and the remaining three substitutions altered the charge or polarity of the native residue. These lesions were spaced throughout the F1 region, and all but one of the mutant proteins (Aer-I123N) were either functional at higher induction levels or rescued in the presence of the high-abundance Tar receptor. Three of the substitutions probably inhibited protein maturation, as these mutant proteins were rapidly proteolyzed, with the exception of a small fraction that was stable and must have folded successfully. A previous study found seven mutations at six sites in the F1 region, all of which reduced the steady-state accumulation of mutant Aer proteins (11). Since Aer maturation requires the proper folding of the protein prior to its export from GroEL (19) and Aer protein folding and stability depend largely on PAS-HAMP interaction (19, 41), the F1 region may be important for aligning these domains. Given the tolerance of this region to substitution, we conclude that the F1 region is not intimately involved in Aer signal transduction but probably provides structural stability to the PAS and/or HAMP domains.
Structural relationships of F1.
Since the functional unit of Aer is a homodimer, it was possible that the F1 domains form a dimeric interface like the AS-2 helices of the HAMP domain. However, stepwise testing for disulfide formation at each Cys substitution in the F1 domain showed sparse cross-linking, indicating that these regions do not form an interactive face (Fig. 3A). One notable region that formed disulfide bonds resided in the central portion of F1, where three residues (A135C, G136C, and R137C) appeared to form a loop (Fig. 3A). When a molecular ruler capable of cross-linking Cys residues within a 6- to 10-Å range was employed, cognate cysteines cross-linked at approximately one-third of the F1 positions (Fig. 3B). The pattern of residues that were accessible and inaccessible to BMOE was consistent with a secondary structure consisting of a proximal helix followed by an extended loop at residues 134 to 139. This pattern supports the secondary structure predicted for the region (Fig. 3). However, there was sparse cross-linking in the C-terminal region and no identifiable features emerged to support the structure predicted in the SYMPRED analysis, except for a short loop at residues 150 to 152.
We investigated the orientation of the F1 region within a trimer of dimers by employing the trifunctional thio-reactive cross-linker TMEA. With the most accessible Cys residue (R137C) as bait, the probe trapped three monomers. The simplest explanation for this is that F1 regions from three different dimers face inward within a trimer of dimers. We further evaluated whether F1-F1′ disulfide cross-linking occurred between or within dimers by titrating with the Tar receptor and measuring the extent of cross-linking. Tar diminished cross-linking (and therefore collisions) between F1 regions, arguing that F1 contacts are between rather than within dimers. The types of arrangement that could account for these data are limited, as the HAMP domains are at the dimer interface and therefore centrally positioned (discussed below).
Homology model of Aer-F1.
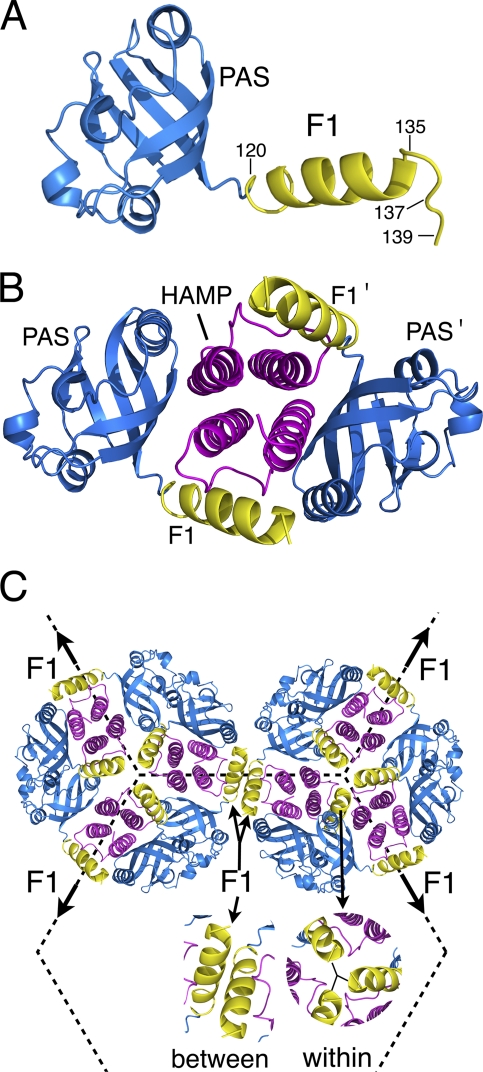
The soluble sensor MmoS from M. capsulatus (Bath) has two FAD binding PAS domains linked by an extended helix with sequence similarity to the N-terminal F1 region of Aer (see Fig. S1 in the supplemental material). Recently, the crystal structure of both PAS domains and the linker was resolved (36), and we used the MmoS structures to create a homology model of the Aer PAS and N-terminal F1 regions. As shown in Fig. 6A, the model of F1 has an extended helix followed by a loop, consistent with the Aer cross-linking data (Fig. 3B and C). From this homology model, we oriented the Aer monomers in a way consistent with the present and previous data.
FIG. 6.
Homology models of the Aer PAS, F1, and HAMP domains oriented in positions consistent with the present and previous data. (A) Ribbon model of the Aer PAS (residues 20 to 119) and the proximal region of F1 (residues 120 to 139) based on the redox sensor domain of MmoS from M. capsulatus (36). The structure of F1 is consistent with the cross-linking data from this study (Fig. 3). (B) Two Aer PAS/F1 domains are positioned beside an Aer-HAMP homology model based on AF1503 from Archeoglobus fulgidus (20). The PAS β scaffolding is proposed to face inward toward the HAMP domains (12). (C) Six dimers of Aer are positioned on a hexagonal grid spaced 12 nm from center to center (10). Three F1 regions were cross-linked with the trigonal probe TMEA, but the proximity of neighboring trimers of dimers suggests that F1 regions may also cross-link between trimers of dimers. Under native, low accumulation levels, Aer would primarily form mixed trimers of dimers with methyl-accepting chemotaxis proteins (16).
Since the Aer PAS and HAMP domains interact (12) and HAMP domains (including that of Aer) dimerize as a four-helix bundle (20; K. Watts, unpublished data), the only symmetric model is one in which the HAMP domains lie between the two PAS domains. From the location of “signal-on” lesions in the Aer PAS domain and data from PAS-HAMP cross-linking, the β strands of the PAS domain scaffold are hypothesized to face inward toward the HAMP domains (12). With these restrictions, the F1 regions in the homology model would project inward such that the PAS and F1 segments circumscribe the HAMP domain (Fig. 6B). The dimensions of this region of the Aer receptor, parallel to the cytoplasmic membrane, would be approximately 67 by 35 Å (Fig. 6B). When positioned onto the hexagonal geometry common to all trimer-of-dimer units at the CheA/W scaffolding of chemoreceptors (10), the possible arrangements of the Aer dimers are limited. Trimers of receptor dimers form a hexagonal (honeycomb) arrangement where the distance between the centers of the hexagons is 12 nm (10), resulting in a spacing between adjacent trimers of dimers of approximately 6.9 nm. Using these constraints, we tested the orientations of Aer dimers in a trimer of dimers and found that the most spatially conservative orientation allowed F1 regions from three monomers to contact within a trimer of dimers (Fig. 6C). Moreover, depending on the angular arrangement between trimers of dimers, the F1 regions between trimers of dimers were also in a position to collide (Fig. 6C). This positioning is consistent with the data from both the present and previous studies. The placement of F1 within a trimer of dimers is consistent with the trigonal cross-linking by TMEA and the decrease in cross-linking as Tar was titrated (Fig. 4). Collisions between trimers of dimers could explain the previous counterintuitive finding that R137C formed multimers when paired with either inter- or intradimeric cross-linked residues (3). The lateral distances between the F1 regions modeled in Fig. 6C indicate a plausible explanation, i.e., that R137C and other exclusively interdimeric cross-linkers may cross-link with monomers from two different dimers (Fig. 6C).
Under native chromosomal expression, most Aer dimers would be found in mixed trimers of dimers with other chemoreceptors. However, we propose that the relative orientation of Aer F1 regions will be the same in either mixed or homogeneous trimers of dimers. This is based on the finding that the kinase control module rather than the PAS domain or membrane region limits the relative orientation of Aer dimers within a trimer of dimers (4). Apart from the influence of the arginine residues at the membrane-cytosol interface, the most likely role of F1 is to assist the orientation between the Aer PAS and HAMP domains. Given the low sequence conservation, as well as the tolerance to substitutions, the guidance is not precise and there must be some dynamic freedom allowed for the PAS and HAMP contact regions to associate.
Supplementary Material
Acknowledgments
We are grateful to Nathan Abraham for technical assistance, Claudia Studdert for the Tsr antiserum, and John S. Parkinson for wild-type Tar expression plasmid pLC113.
This work was supported by a grant from the National Institute of General Medical Sciences (GM29481) to B. L. Taylor.
Footnotes
Published ahead of print on 19 November 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Ames, P., and J. S. Parkinson. 2006. Conformational suppression of inter-receptor signaling defects. Proc. Natl. Acad. Sci. U. S. A. 103:9292-9297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ames, P., C. A. Studdert, R. H. Reiser, and J. S. Parkinson. 2002. Collaborative signaling by mixed chemoreceptor teams in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 99:7060-7065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amin, D. N. 2006. Membrane organization and multimeric interactions of the Aer receptor in E. coli. Ph.D. dissertation. Loma Linda University, Loma Linda, CA.
- 4.Amin, D. N., B. L. Taylor, and M. S. Johnson. 2007. Organization of the aerotaxis receptor Aer in the membrane of Escherichia coli. J. Bacteriol. 189:7206-7212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amin, D. N., B. L. Taylor, and M. S. Johnson. 2006. Topology and boundaries of the aerotaxis receptor Aer in the membrane of Escherichia coli. J. Bacteriol. 188:894-901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bass, R. B., S. L. Butler, S. A. Chervitz, S. L. Gloor, and J. J. Falke. 2007. Use of site-directed cysteine and disulfide chemistry to probe protein structure and dynamics: applications to soluble and transmembrane receptors of bacterial chemotaxis. Methods Enzymol. 423:25-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bertani, G. 2004. Lysogeny at mid-twentieth century: P1, P2, and other experimental systems. J. Bacteriol. 186:595-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bibikov, S. I., L. A. Barnes, Y. Gitin, and J. S. Parkinson. 2000. Domain organization and flavin adenine dinucleotide-binding determinants in the aerotaxis signal transducer Aer of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 97:5830-5835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bibikov, S. I., R. Biran, K. E. Rudd, and J. S. Parkinson. 1997. A signal transducer for aerotaxis in Escherichia coli. J. Bacteriol. 179:4075-4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Briegel, A., et al. 2009. Universal architecture of bacterial chemoreceptor arrays. Proc. Natl. Acad. Sci. U. S. A. 106:17181-17186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burón-Barral, M. C., K. K. Gosink, and J. S. Parkinson. 2006. Loss- and gain-of-function mutations in the F1-HAMP region of the Escherichia coli aerotaxis transducer Aer. J. Bacteriol. 188:3477-3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Campbell, A. J., K. J. Watts, M. S. Johnson, and B. L. Taylor. 2010. Gain-of-function mutations cluster in distinct regions associated with the signalling pathway in the PAS domain of the aerotaxis receptor, Aer. Mol. Microbiol. 77:575-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang, A. C., and S. N. Cohen. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 134:1141-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cuff, J. A., and G. J. Barton. 2000. Application of multiple sequence alignment profiles to improve protein secondary structure prediction. Proteins 40:502-511. [DOI] [PubMed] [Google Scholar]
- 15.Edwards, J. C., M. S. Johnson, and B. L. Taylor. 2006. Differentiation between electron transport sensing and proton motive force sensing by the Aer and Tsr receptors for aerotaxis. Mol. Microbiol. 62:823-837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gosink, K. K., M. del Carmen Buron-Barral, and J. S. Parkinson. 2006. Signaling interactions between the aerotaxis transducer Aer and heterologous chemoreceptors in Escherichia coli. J. Bacteriol. 188:3487-3493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Green, N. S., E. Reisler, and K. N. Houk. 2001. Quantitative evaluation of the lengths of homobifunctional protein cross-linking reagents used as molecular rulers. Protein Sci. 10:1293-1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hazelbauer, G. L., J. J. Falke, and J. S. Parkinson. 2008. Bacterial chemoreceptors: high-performance signaling in networked arrays. Trends Biochem. Sci. 33:9-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herrmann, S., Q. Ma, M. S. Johnson, A. V. Repik, and B. L. Taylor. 2004. PAS domain of the Aer redox sensor requires C-terminal residues for native-fold formation and flavin adenine dinucleotide binding. J. Bacteriol. 186:6782-6791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hulko, M., et al. 2006. The HAMP domain structure implies helix rotation in transmembrane signaling. Cell 126:929-940. [DOI] [PubMed] [Google Scholar]
- 21.Jones, D. T. 1999. Protein structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292:195-202. [DOI] [PubMed] [Google Scholar]
- 22.Jung, K. H., and J. L. Spudich. 1998. Suppressor mutation analysis of the sensory rhodopsin I-transducer complex: insights into the color-sensing mechanism. J. Bacteriol. 180:2033-2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Larkin, M. A., et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947-2948. [DOI] [PubMed] [Google Scholar]
- 24.Lee, E., and C. Manoil. 1994. Mutations eliminating the protein export function of a membrane-spanning sequence. J. Biol. Chem. 269:28822-28828. [PubMed] [Google Scholar]
- 25.Lin, K., V. A. Simossis, W. R. Taylor, and J. Heringa. 2005. A simple and fast secondary structure prediction method using hidden neural networks. Bioinformatics 21:152-159. [DOI] [PubMed] [Google Scholar]
- 26.Ma, Q., F. Roy, S. Herrmann, B. L. Taylor, and M. S. Johnson. 2004. The Aer protein of Escherichia coli forms a homodimer independent of the signaling domain and flavin adenine dinucleotide binding. J. Bacteriol. 186:7456-7459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parkinson, J. S. 1978. Complementation analysis and deletion mapping of Escherichia coli mutants defective in chemotaxis. J. Bacteriol. 135:45-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pollastri, G., D. Przybylski, B. Rost, and P. Baldi. 2002. Improving the prediction of protein secondary structure in three and eight classes using recurrent neural networks and profiles. Proteins 47:228-235. [DOI] [PubMed] [Google Scholar]
- 29.Rebbapragada, A., et al. 1997. The Aer protein and the serine chemoreceptor Tsr independently sense intracellular energy levels and transduce oxygen, redox, and energy signals for Escherichia coli behavior. Proc. Natl. Acad. Sci. U. S. A. 94:10541-10546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Repik, A., et al. 2000. PAS domain residues involved in signal transduction by the Aer redox sensor of Escherichia coli. Mol. Microbiol. 36:806-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rost, B., and C. Sander. 1993. Improved prediction of protein secondary structure by use of sequence profiles and neural networks. Proc. Natl. Acad. Sci. U. S. A. 90:7558-7562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Studdert, C. A., and J. S. Parkinson. 2004. Crosslinking snapshots of bacterial chemoreceptor squads. Proc. Natl. Acad. Sci. U. S. A. 101:2117-2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taylor, B. L. 2007. Aer on the inside looking out: paradigm for a PAS-HAMP role in sensing oxygen, redox and energy. Mol. Microbiol. 65:1415-1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Taylor, B. L., M. S. Johnson, and K. J. Watts. 2003. Signal transduction in prokaryotic PAS domains, p. 15-50. In S. T. Crews (ed.), PAS proteins: regulators and sensors of development and physiology. Kluwer Academic Publishers, Norwell, MA.
- 35.Taylor, B. L., K. J. Watts, and M. S. Johnson. 2007. Oxygen and redox sensing by two-component systems that regulate behavioral responses: behavioral assays and structural studies of aer using in vivo disulfide cross-linking. Methods Enzymol. 422:190-232. [DOI] [PubMed] [Google Scholar]
- 36.Ukaegbu, U. E., and A. C. Rosenzweig. 2009. Structure of the redox sensor domain of Methylococcus capsulatus (Bath) MmoS. Biochemistry 48:2207-2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vanhercke, T., C. Ampe, L. Tirry, and P. Denolf. 2005. Reducing mutational bias in random protein libraries. Anal. Biochem. 339:9-14. [DOI] [PubMed] [Google Scholar]
- 38.von Heijne, G. 1994. Membrane proteins: from sequence to structure. Annu. Rev. Biophys. Biomol. Struct. 23:167-192. [DOI] [PubMed] [Google Scholar]
- 39.Watts, K. J., M. S. Johnson, and B. L. Taylor. 2006. Minimal requirements for oxygen sensing by the aerotaxis receptor Aer. Mol. Microbiol. 59:1317-1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Watts, K. J., M. S. Johnson, and B. L. Taylor. 2008. Structure-function relationships in the HAMP and proximal signaling domains of the aerotaxis receptor Aer. J. Bacteriol. 190:2118-2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Watts, K. J., Q. Ma, M. S. Johnson, and B. L. Taylor. 2004. Interactions between the PAS and HAMP domains of the Escherichia coli aerotaxis receptor Aer. J. Bacteriol. 186:7440-7449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Watts, K. J., K. Sommer, S. L. Fry, M. S. Johnson, and B. L. Taylor. 2006. Function of the N-terminal cap of the PAS domain in signaling by the aerotaxis receptor Aer. J. Bacteriol. 188:2154-2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu, H. S., et al. 2002. Aerotactic responses in bacteria to photoreleased oxygen. FEMS Microbiol. Lett. 217:237-242. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.