FIG. 1.
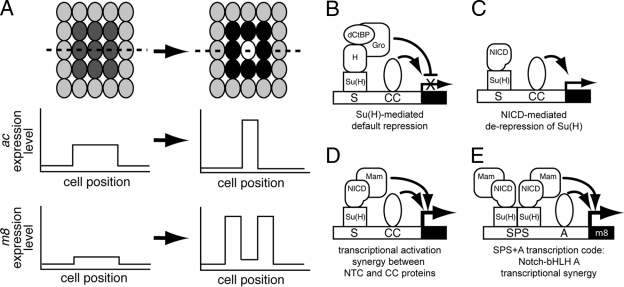
Proneural ac and E(spl)-C m8 gene expression patterns during Notch signaling mediated lateral inhibition in Drosophila proneural clusters. (A) In the early proneural cluster, both the model proneural and E(spl)-C genes, ac and m8, respectively, are expressed uniformly at low levels. In the late proneural cluster, after Notch signaling is activated, m8 is transcribed at high levels in the nonneural precursor cells (non-NPCs [black cells]). In contrast, at these later stages, ac is strongly expressed in the neural precursor cell (NPC [white cell]). The relative m8 and ac gene expression levels along the broken line bisecting the proneural cluster are shown below the proneural clusters. (B to E) Current models for Su(H)-regulated transcription. (B) In the absence of active Notch signaling and NICD, expression of target genes is blocked by Su(H)-mediated default repression. In this situation, corepressor proteins, such as Hairless (H), Groucho (Gro), and dCtBP, bind Su(H) and repress gene transcription. (C) When Notch signaling is activated, the cleaved Notch intracellular domain (NICD) translocates to the nucleus and displaces corepressor protein complexes bound to Su(H). Formation of the Su(H)/NICD binary complex and displacement of corepressors results in NICD-mediated derepression of Su(H). (D) The binary complex can recruit coactivators, such as Mastermind (Mam), and the resulting ternary complex can synergistically interact with other transcription factors (combinatorial cofactors [CC]) also bound nearby on the DNA. (E) On the model E(spl)-C promoter, m8, the SPS+A transcription code mediates synergistic interactions between Notch transcription complexes (NTC) and bHLH A combinatorial cofactors that strongly upregulate m8 expression in the non-NPCs.