FIG. 1.
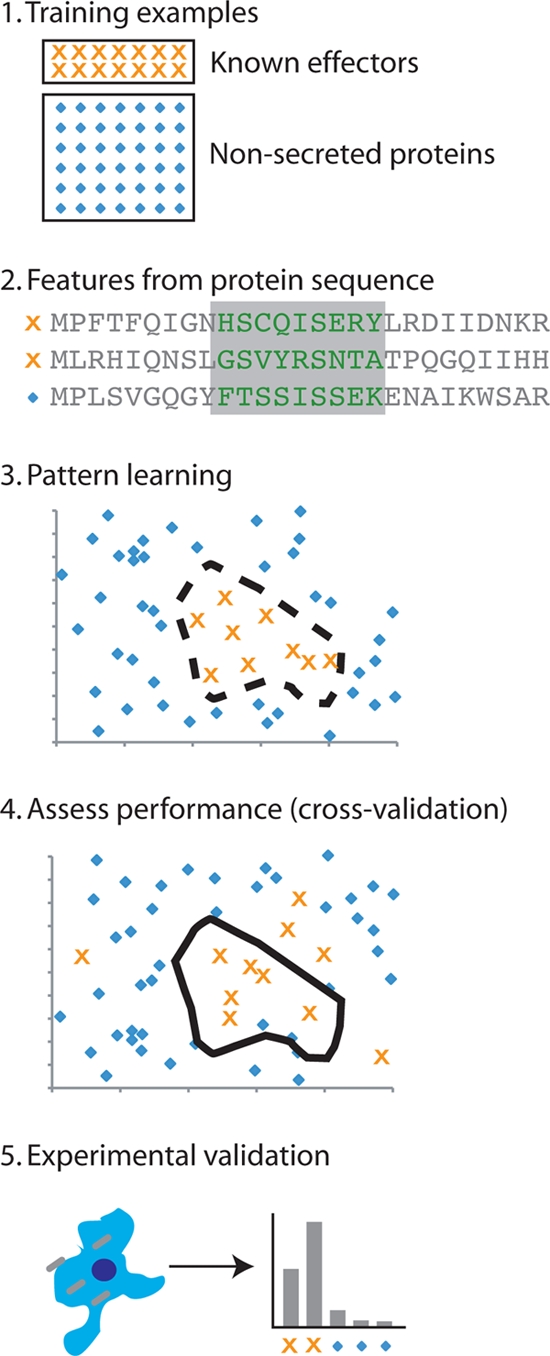
Machine learning approaches to secreted effector identification. Each of the methods described in this review follows a similar process. Step 1, sets of known secreted effectors (positive examples) and proteins that are not secreted or assumed to be not secreted (negative examples) are chosen. Step 2, features of the protein sequence (e.g., amino acid conservation, sequence, phylogenetic distribution, etc.) are derived from all proteins and transformed into a numeric representation. Step 3, a machine learning algorithm (e.g., a support vector machine) learns to discriminate the positive examples from negative examples in a high-dimensional space formed by the chosen protein features. Step 4, the performance of the approach is assessed by applying the model learned in step 3 to independent examples that were not included in the training. Step 5, experimental validation must then be applied to finally determine whether or not a protein is secreted.
