Abstract
Burkholderia cenocepacia is a significant opportunistic pathogen in individuals with cystic fibrosis. ShvR, a LysR-type transcriptional regulator, has previously been shown to influence colony morphology, biofilm formation, virulence in plant and animal infection models, and some quorum-sensing-dependent phenotypes. In the present study, it was shown that ShvR negatively regulates its own expression, as is typical for LysR-type regulators. The production of quorum-sensing signal molecules was detected earlier in growth in the shvR mutant than in the wild type, and ShvR repressed expression of the quorum-sensing regulatory genes cepIR and cciIR. Microarray analysis and transcriptional fusions revealed that ShvR regulated over 1,000 genes, including the zinc metalloproteases zmpA and zmpB. The shvR mutant displayed increased gene expression of the type II secretion system and significantly increased protease and lipase activities. Both ShvR and CepR influence expression of a 24-kb genomic region adjacent to shvR that includes the afcA and afcC operons, required for the production of an antifungal agent; however, the reduction in expression was substantially greater in the shvR mutant than in the cepR mutant. Only the shvR mutation resulted in reduced antifungal activity against Rhizoctonia solani. ShvR, but not CepR, was shown to directly regulate expression of the afcA and afcC promoters. In summary, ShvR was determined to have a significant influence on the expression of quorum-sensing, protease, lipase, type II secretion, and afc genes.
Members of the Burkholderia cepacia complex (Bcc) are important in medical, agricultural, and biotechnological research fields (31, 35). Seventeen Bcc species have been identified, including Burkholderia cenocepacia, which is an opportunistic pathogen in individuals with the inherited disease cystic fibrosis (CF). B. cenocepacia is intrinsically resistant to antibiotics and can persist in the lungs of CF patients (35, 44). In some patients, infection with B. cenocepacia can progress from chronic lung infection to what is termed “cepacia syndrome.” Cepacia syndrome is associated with a rapid deterioration in lung function associated with necrotizing pneumonia, bacteremia, and sepsis that can result in death (24).
Our laboratory previously identified spontaneous shiny colony variants (shv) that were easily distinguished from the typical rough colony morphotypes of B. cenocepacia (4). These shv typically exhibited the absence of an extracellular matrix, reduced biofilm formation, and reduced virulence in alfalfa and rat infection models and displayed differences in N-acyl homoserine lactone (AHL) and protease activities, motility, and siderophore production. Expression of BCAS0225, encoding the LysR-type transcriptional regulator (LTTR) ShvR, influenced a number of these phenotypes in shv (4).
LTTRs are part of a large protein family and display a well-conserved structure with an N-terminal DNA-binding helix-turn-helix motif and a C-terminal coinducer-binding domain (for a review, see reference 33). LTTRs typically engage in negative autoregulation and frequently positively regulate divergently transcribed genes. However, it is now widely accepted that LTTRs can act as global regulators in a positive or negative manner (33).
The locus carrying shvR lies adjacent to the predicted afcA and afcC operons that are divergently transcribed (60). Mutations affecting afcA, afcC, and afcD in Burkholderia pyrrocinia BC11 (formerly B. cepacia BC11 [56]) were previously shown to impair the production of an antifungal compound and inhibit antifungal activity against the soilborne phytopathogen Rhizoctonia solani (25). We previously showed that the 27-kb genomic region encompassing shvR and the afc operons was influenced by quorum sensing (QS). Expression of shvR and many genes in the afcA and afcC operons was positively regulated by CepR and negatively regulated by CciR (41). QS is a cell-cell communication system involving the production and perception of chemical signals used by diverse bacterial species. In many Gram-negative bacteria, members of the LuxI protein family synthesize AHLs that bind and activate cognate LuxR protein family transcriptional regulators (for reviews, see references 40, 49, and 57). AHLs accumulate during bacterial growth until a threshold is reached at high cell densities, allowing coordinated regulation of gene expression.
B. cenocepacia has two complete AHL-dependent QS systems as well as an orphan LuxR homolog. CepI is primarily responsible for the synthesis of N-octanoyl-l-homoserine lactone (OHL) (29) and minor amounts of N-hexanoyl-l-homoserine lactone (HHL) (30). CciI primarily synthesizes HHL, with lesser amounts of OHL produced (36). All members of the Bcc have the CepIR system (19, 32), whereas CciIR is found only in transmissible B. cenocepacia strains containing the cenocepacia island and is absent from genomes of other B. cenocepacia strains, including representatives of the transmissible PHDC lineage (AU1054, HI2424) (2, 21, 60). CepR2 is an orphan LuxR homolog found in all B. cenocepacia strains and can regulate gene expression in the absence of AHLs (37). The B. cenocepacia CepIR system positively influences virulence in murine, alfalfa, Caenorhabditis elegans, Galleria mellonella, and Danio rerio infection models (22, 26, 27, 51, 55, 58). A B. cenocepacia cciI mutant exhibited reduced virulence in a rat chronic respiratory infection model but did not affect virulence in Caenorhabditis elegans, Galleria mellonella, or alfalfa infection models (2, 55). CepR, CciR, and CepR2 regulate between 3 and 12% of the genome and thus are considered global regulators (37, 41). CepIR and CciIR regulate protease and lipase activities and type II, III, and VI secretion systems, as well as biofilm formation and maturation (8, 23, 26, 29, 36, 41, 51, 53, 54).
We previously demonstrated an association between shv and the shvR mutation with some QS-dependent phenotypes (4). To further investigate this relationship, we studied the effects of ShvR on the AHL-dependent QS regulatory network and examined the coregulation of gene expression by CepR, CciR, and ShvR. We investigated the influence of ShvR on several QS-controlled phenotypes.
MATERIALS AND METHODS
Strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Cultures were routinely grown at 37°C in Miller's Luria broth (LB) (Invitrogen, Burlington, Ontario, Canada) with shaking or on 1.5% Lennox LB agar plates. For promoter::lux assays, strains were grown in LB, Trypticase soy broth (TSB) (Difco, Franklin Lakes, NJ), 0.25% Trypticase soy broth with 5% Bacto peptone (Difco) (PTSB), dialyzed brain heart infusion (D-BHI), and/or 1.5% skim milk (Difco). For some assays, OHL was added in concentrations ranging from 30 to 3,000 pM. For microarray and quantitative reverse transcription-PCR (qRT-PCR) experiments, cultures were grown to stationary phase (16 h) in 10 ml LB in 125-ml Erlenmeyer flasks. For promoter::lux assays, samples were cultured in black, clear-bottom, 96-well plates (Corning, Inc., Corning, NY). Cultures entered stationary phase at 18 h (growing at 37°C) or 30 h (growing at 29°C). AHL activity was monitored using Agrobacterium tumefaciens A136(pCF218)(pMV26) in a real-time liquid coculture assay with B. cenocepacia grown in 96-well plates in TSB at 29°C as previously described (3). Protease activity was determined using D-BHI with 1.5% skim milk agar plates as previously described (50) except that cell pellets were washed twice with PTSB and normalized to an optical density (OD) of 0.3 prior to inoculation. Lipase activity was determined as previously described (23) except that cell pellets were washed twice with LB and normalized to an OD of 0.3 prior to inoculation on 1% polysorbate (Tween) 80 or 1% tributyrin in LB agar plates. Colony morphology was assessed by examining the appearance of rough or shiny morphology of strains on LB agar following incubation at 28°C or 37°C for 72 h and capturing images using a Samsung digital camera. Biofilm formation was assessed as previously described (5) on polystyrene pegs in a 96-well-plate format (Nunc, Roskilde, Denmark) by using cultures grown in TSB for 24 h at 37°C on a rocking platform. Biomass was strained with crystal violet, destained with ethanol, and quantified by measuring OD at 600 nm. When appropriate, the following concentrations of antibiotics were used: 100 μg/ml of trimethoprim (Tp) and 200 μg/ml of tetracycline (Tc) for B. cenocepacia, 100 μg/ml of Tp and 50 μg/ml of kanamycin (Km) for Escherichia coli, and 25 μg/ml of Km and 4.5 μg/ml of Tc for Agrobacterium tumefaciens. Antibiotics were purchased from Sigma-Aldrich Canada Ltd. (Oakville, Ontario, Canada).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Reference |
|---|---|---|
| Strains | ||
| A. tumefaciens | ||
| A136 | Ti plasmidless host | C. Fuqua |
| E. coli | ||
| TOP10 | F−mcrAΔ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 deoR recA1 araD139 Δ(ara-leu)7697 galU galK rpsL (Str) endA1 nupG | Invitrogen |
| DH10B | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 recA1 endA1 araD139 Δ(ara-leu)7697 galU galK λ−rpsL nupG | Invitrogen |
| SY327 | araD Δ(lac-pro) argE(Am) recA56 RifrnalA λpir | 39 |
| B. cenocepacia | ||
| K56-2 | CF isolate | 34 |
| K56-R2 | cepR::Tn5-OT182 derivative of K56-2, Tcr | 29 |
| K56-2ΔcepR | cepR derivative of K56-2 | 41 |
| K56-2shvR::Tp | shvR::Tp derivative of K56-2, Tpr | This study |
| K56-2ΔshvR | shvR derivative of K56-2 | This study |
| K56-2cepIcciIb | ΔcepI cciI::Tp derivative of K56-2, Tpr | 36 |
| Plasmids | ||
| pCR2.1Topo | Cloning vector for PCR products, Apr Kmr | Invitrogen |
| pCF218 | IncP plasmid expressing TraR, Tcr | 63 |
| pMV26 | traI-luxCDABE fusion, Kmr | 51 |
| pRK2013 | ColEl Tra (RK2)+, Kmr | 15 |
| pGSVTplux | Mobilizable suicide vector containing lux operon, derivative from pGSV3-lux, OriT, Tpr | 4 |
| pEX18Tc | Gene replacement vector oriT+, sacB+, Tcr | 20 |
| pUCP26-BCAS0225 | pUCP26 with 1.7-kb PstI-BamHI fragment containing BCAS0225 and upstream region, Tcr | 4 |
| pEX18Tc-BCAS0225::Tp | pEX18Tc containing BCA0225 inactivated with a Tp cassette from pGSVTplux | This study |
| pGPI-SceI | oriR6K, Tpr, mob+, carries I-SceI endonuclease recognition site, Tpr | 16 |
| pDAI-SceI | pDA17 carrying the I-SceI gene, Tcr | 16 |
| pUCP28T | Broad-host-range vector, Tpr | 48 |
| p28T-shvR | pUCP28T with 1.7-kb PstI-BamHI fragment containing BCAS0225 and upstream region, Tcr | This study |
| pSLR100 | pUCP28T with 1.65-kb KpnI-SphI fragment from pSLA3.2 containing cepR gene, Tpr | 29 |
| pMS402 | lux-based promoter reporter plasmid, Kmr Tpr | 13 |
| pCP300 | cepI::lux transcriptional fusion constructed in pMS402, Kmr Tpr | 36 |
| pRM432 | cepR::lux transcriptional fusion constructed in pMS402, Kmr Tpr | 36 |
| pRM445 | cciIR::lux transcriptional fusion constructed in pMS402, Kmr Tpr | 36 |
| pPafcA | afcA::lux transcriptional fusion constructed in pMS402, Kmr Tpr | 41 |
| pPafcC | afcC::lux transcriptional fusion constructed in pMS402, Kmr Tpr | This study |
| pBS13 | zmpA::lux transcriptional fusion constructed in pMS402, Kmr Tpr Tcr | 37 |
| pBS9 | zmpB::lux transcriptional fusion constructed in pMS402, Kmr Tpr Tcr | 26 |
| pPgspC | gspC::lux transcriptional fusion constructed in pMS402, Kmr Tpr | This study |
| pPgspG | gspG::lux transcriptional fusion constructed in pMS402, Kmr Tpr | This study |
| pPshvR | shvR::lux transcriptional fusion constructed in pMS402, Kmr Tpr | This study |
| pCS26-Pac | lux-based promoter reporter plasmid, Kmr | 6 |
| pEPO100 | pCS26-Pac with 269-bp XhoI-BamHI fragment from pCP300 containing cepI promoter, Kmr | This study |
| pEPO101 | pCS26-Pac with 371-bp XhoI-BamHI fragment from pPafcA containing afcA promoter, Kmr | This study |
| pEPO128 | pCS26-Pac with 405-bp XhoI-BamHI fragment from pPafcC containing afcC promoter, Kmr | This study |
Antifungal activity.
Antagonistic activities against Rhizoctonia solani (obtained from the culture collection of the phytopathology group of the Institute of Plant Sciences, Federal Institute of Technology, Zurich, Switzerland) were assayed on malt extract agar (15 g/liter; supplemented with 12 g/liter agar; Becton Dickinson, Sparks, MD) by spotting 10-μl stationary-phase cultures of selected strains at three positions on the plate. Following incubation overnight at 37°C, a 5-mm-diameter fungal inoculum, which was cut from a 1-week-old fungal culture plate, was placed onto the center of the plate. Plates were incubated at 22°C in the dark, and inhibition zones were recorded after 3 days. The experiment was performed in triplicate.
DNA manipulations.
DNA manipulations were performed using standard techniques as described previously (45), and genomic DNA was isolated as described previously (1). Oligonucleotide primers (Table 2) were designed with Primer3 (42) and were synthesized by the University of Calgary Core DNA and Protein Services (Calgary, Alberta, Canada). Plasmids were introduced into B. cenocepacia by electroporation (12).
TABLE 2.
Oligonucleotide primers used in this study
| Primer | Sequence (5′→3′) | Reference |
|---|---|---|
| 5′-XbaI-UP-BCAS0225 | TTTTTCTAGATGCGCATCGAATGCACACCG | This study |
| 3′-KpnI-UP-BCAS0225 | TTTTGGTACCCTTCTCAATCGCTTTGCC | This study |
| 5′-KpnI-DWN-BCAS0225 | TTTTGGTACCACGTGCTGCCGATGTATCGC | This study |
| 3′-SmaI-DWN-BCAS0225 | TTTTCCCGGGATCTTCCAGATTCACGTCCG | This study |
| XhoI-5′-PBCAS0225 | AGCTCTCGAGCGGATTCATCTTGACGGTCG | This study |
| PBCAS0225-3′-BamHI | AGCTGGATCCGGCAGACTTCTCAATCGCTT | This study |
| PgspC&Gfor1 | GGCTCGAGTGTCGAGACGATACAGCTTGAG | This study |
| PgspC&Grev1 | GGGGATCCCATATTTGCCATCCATTTGC | This study |
| PafcCfor1 | GGCTCGAGCGATTCGATCAGACGGTGAATG | This study |
| PafcCrev1 | GGGGATCCGACAAGCGATGAGGTGACG | This study |
| Fndh | GCGATCGGGCTGTACAAGTT | 53 |
| Rndh | AGTGGCTCAGCGACTGGAA | 53 |
| BCAS0220qRTfor2 | GAACCAGTTCTCGGTGTTCG | This study |
| BCAS0220qRTrev2 | GATCCAGTTGCTCATCGACA | This study |
| BCAS0215qRTfor1 | ATCCTGTCGATGCTGCTGAT | This study |
| BCAS0215qRTrev1 | TACGAGCAGGTCATCCAGTC | This study |
| BCAS0208qRTfor1 | ATCTGCAAGGCGTTCATCTC | This study |
| BCAS0208qRTrev1 | GTAGTTCGTGCCTTCCCAGA | This study |
| BCAS0204qRTfor1 | GCAATTGCAGAAGGTCGAGT | This study |
| BCAS0204qRTrev1 | CGACGATCTCGGATACACG | This study |
| pUCP28TflankMCSfor1 | CGACGTTGTAAAACGACGG | This study |
| pUCP28TflankMCSrev1 | GGAAACAGCTATGACCATGA | This study |
Construction of the K56-2shvR::Tp and K56-2ΔshvR mutants.
To construct K56-2shvR::Tp, pUCP26-BCAS0225 (4) and pEX18Tc (20) were digested with PstI and EcoRI, respectively. The resulting insert or vector fragments were blunt ended using T4 DNA polymerase (Invitrogen), digested with BamHI, and ligated to create pEX18Tc-BCAS0225. BCAS0225 was inactivated using a Tp cassette from pGSVTplux (4) by using SacI to generate pEX18Tc-BCAS0225::Tp. The K56-2shvR::Tp mutant was generated by gene replacement by using the previously described strategy (47). A K56-2ΔshvR deletion mutant was constructed by following a described method (16) by using the primers listed in Table 2. PCR confirmed gene replacement or deletion in these mutants.
Transcriptional fusions to luxCDABE (lux).
Promoter regions for shvR, afcC, gspC, and gspG were amplified using the described primers (Table 2) and cloned into the XhoI-BamHI site upstream of lux in pMS402 (13) or pCS26-Pac (6). For the expression reporter system, cepR or shvR expression constructs in vector pUCP28T were electroporated into pCS26-Pac derivatives, and construct presence was confirmed by PCR by using the appropriate primers (Table 2). Luminescence assays were carried out as previously described (8, 41). Cultures for luminescence assays on skim milk agar were prepared as described above, and promoter::lux activity was visualized using a Molecular Imager ChemiDoc system (Bio-Rad, Mississauga, Ontario, Canada).
RNA manipulations.
RNA was prepared as previously described (41) using a RiboPure bacterial RNA isolation kit (Ambion, Streetsville, Ontario, Canada). DNase treatment was performed, and samples were confirmed by PCR using Taq polymerase (Invitrogen) to be free of DNA prior to cDNA synthesis.
Microarray sample preparation.
Microarray samples were prepared as previously described (41). Briefly, three independent RNA samples from B. cenocepacia strains grown to stationary phase in LB were used in microarray experiments. Gene expression profiles were generated using custom B. cenocepacia J2315 microarrays (Agilent, Santa Clara, CA) (21, 28). K56-2 and K56-2shvR::Tp cDNA samples were fluorescently labeled with Cy3 and Cy5, respectively. Labeling, hybridization, and scanning were performed by the Mahenthiralingam Laboratory, Cardiff University, Wales.
Microarray data analysis.
Microarray data analysis was performed using GeneSpring GX 7.3.1 software (Agilent). Initial data were preprocessed by employing the enhanced Agilent FE import method, and then per-spot and per-chip normalizations were performed using the Affymetrix FE data normalization recommended for Agilent arrays, eliminating the per-gene normalization step. After filtering to include genes present in one of three samples, followed by filtering on confidence (P < 0.05, t test with Benjamini-Hochberg and false discovery rate postcorrection), the remaining genes were filtered using a 1.5-fold ratio change to identify genes showing increased or decreased expression in K56-2shvR::Tp compared to that in K56-2. Operon prediction was derived from analysis of B. cenocepacia genomes at http://www.burkholderia.com (60).
qRT-PCR.
Quantitative RT-PCR was performed as previously described on samples from B. cenocepacia strains grown to stationary phase in LB (41) except the NADH dehydrogenase gene ndh (BCAM0166) was used as a reference standard as described previously (53). Expression of ndh was not significantly altered according to microarray analysis (data not shown).
Statistical analyses.
Analysis of variance (ANOVA) was performed with GraphPad Prism software (GraphPad Software, San Diego, CA).
Microarray data accession number.
The entire microarray data set has been deposited in the ArrayExpress database (http://www.ebi.ac.uk/arrayexpress) under accession number E-MEXP-2939.
RESULTS
ShvR expression is temperature regulated and negatively autoregulated.
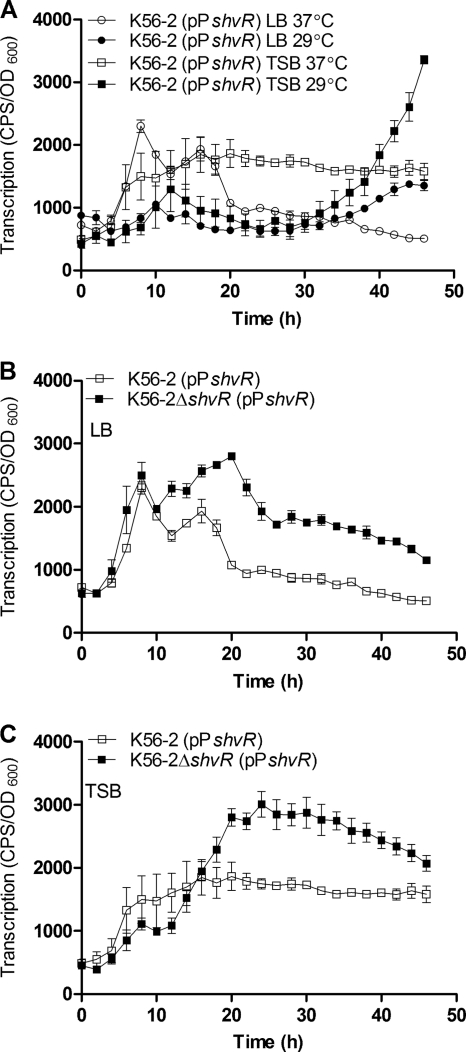
Expression of shvR was monitored using promoter::lux fusions in wild-type cells (Fig. 1 A). In LB or TSB medium incubated at 37°C, shvR::lux expression levels were highest between 8 and 18 h and dropped after this point in LB but remained relatively steady in TSB. During growth in LB or TSB medium at 29°C, the highest shvR::lux expression level occurred after prolonged incubation (Fig. 1A). Negative autoregulation is a common feature of LysR-type regulators (33), which prompted us to examine shvR expression in an shvR mutant versus that in the wild type. Expression of the shvR::lux fusion was significantly higher in the shvR mutant than in K56-2 in both LB and TSB (Fig. 1B and C).
FIG. 1.
Influence of medium and temperature on shvR promoter activity. Expression of an shvR promoter::lux fusion (pPshvR) in K56-2 in LB and TSB media at 29°C or 37°C (A) and in K56-2 and K56-2ΔshvR in LB (B) and TSB (C) at 37°C. Expression was significantly increased in K56-2ΔshvR compared to that in K56-2 from 11 to 38 h in LB and from 22 to 35 h in TSB (P < 0.001, two-way ANOVA). All values are the means ± standard deviations (SD) of results from triplicate cultures and are representative of results from two individual trials.
ShvR temporally affects AHL activity and negatively regulates expression of cepIR and cciIR QS genes.
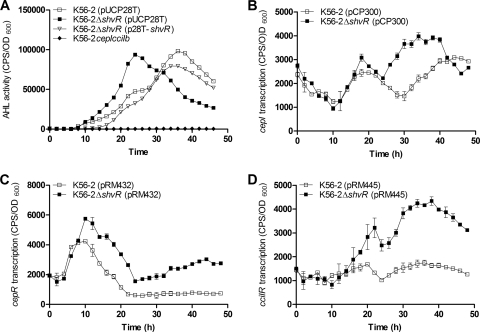
We previously observed that shvR mutants had slightly increased AHL activity compared to that of the wild type (3). AHL activity profiles throughout growth were monitored in a real-time liquid coculture assay with K56-2ΔshvR and K56-2 by using the biosensor Agrobacterium tumefaciens A136(pCF218)(pMV26). AHL activity peaked prior to stationary phase in K56-2ΔshvR(pUCP28T) and during stationary phase in K56-2(pUCP28T), although total AHL activity was similar between the strains (Fig. 2 A). There was no significant difference between K56-2(pUCP28T) and K56-2ΔshvR(p28T-shvR), indicating restoration of parental AHL activity patterns by expression of shvR in trans (Fig. 2A). These data indicate that ShvR affects AHL activity in terms of timing rather than quantity. This trend was confirmed in experiments performed in the absence of vector pUCP28T, where AHL activity peaked prior to stationary phase in two independently constructed shvR mutants compared to that of K56-2 (data not shown).
FIG. 2.
Effect of the shvR mutation on AHL activity and expression of QS genes. (A) AHL activity was measured using the biosensor A. tumefaciens A136(pCF218)(pMV26) in coculture with B. cenocepacia at 29°C. AHL activity was significantly increased from 22 to 26.5 h (log phase) and significantly decreased from 35 to 45 h (stationary phase) in K56-2ΔshvR(pUCP28T) compared to that in K56-2(pUCP28T) (P < 0.05, two-way ANOVA). Expression of cepI (B), cepR (C), or cciIR (D) was monitored using promoter::lux fusions (pCP300, pRM432, or pRM445, respectively) in K56-2 and K56-2ΔshvR in LB at 37°C. Expression is significantly increased in K56-2ΔshvR compared to that in K56-2 from 24.5 to 40.5 h for cepI, from 5.5 to 49.5 h for cepR, and from 19 to 49.5 h for cciIR (P < 0.001, two-way ANOVA). All values are the means ± SD of results from triplicate cultures and are representative of results from at least two individual trials.
Promoter::lux fusions to cepI, cepR, and cciIR were used to assess the effect of ShvR on the expression of QS genes. In the shvR mutant compared to K56-2, cepR expression was significantly higher from mid-log phase, and cepI and cciR expression was significantly higher in stationary phase (Fig. 2B to D). Furthermore, using qRT-PCR, we confirmed that expression of cepI, cepR, and cciIR was increased 3.6-, 5.6-, and 12.5-fold, respectively, in the shvR mutant compared to that in K56-2 in stationary phase (Table 2).
ShvR coregulates QS-controlled genes but independently regulates biofilm formation and rough colony morphology.
Because ShvR influences timing of AHL activity and expression of cepIR and cciIR, we compared microarray expression profiles generated from stationary-phase cultures of the cepR, cciR (41), and shvR mutants relative to K56-2. Microarray analysis showed that 1,077 genes were expressed differently in the shvR mutant than in K56-2 (Table 3). Among the coregulated genes, patterns of common and reciprocal regulation were observed. CepR and ShvR commonly regulated 263 genes; 159 genes were positively regulated and 100 genes negatively regulated by CepR and ShvR. Only four genes were reciprocally regulated by CepR and ShvR, whereas most of the 156 genes coregulated by CciR and ShvR were reciprocally regulated (Table 3). Of these reciprocally regulated genes, 135 genes were positively regulated by ShvR and negatively regulated by CciR, and 14 genes were negatively regulated by ShvR and positively regulated by CciR. Seven genes were commonly regulated by ShvR and CciR, including cciI, which was negatively regulated (Table 3). Negative regulation of cciI by ShvR was observed in cciIR::lux fusions, microarray data, and qRT-PCR (Fig. 2D and Table 4). The expression of 369 and 358 genes was decreased and increased, respectively, in the shvR mutant compared to that of K56-2 but unaffected by mutation of cepR or cciR (Table 3). These represent genes independently regulated by ShvR and include genes encoding 18 LTTRs, exopolysaccharide biosynthesis proteins, ABC transporter ATP-binding proteins, 45 membrane proteins, 20 exported proteins, and 15 lipoproteins.
TABLE 3.
Microarray analysis of genes controlled by ShvR, CepR, and CciR
| Regulators | Regulation typee | No. of genes |
|---|---|---|
| ShvR (total no.)a | Positive | 595 |
| Negative | 482 | |
| ShvR (unaffected by cepR or cciR)b | Positive | 369 |
| Negative | 358 | |
| Coregulated by ShvR and CepRc | cepR− shvR− | 100 |
| cepR− shvR+ | 3 | |
| cepR+ shvR− | 1 | |
| cepR+ shvR+ | 159 | |
| Coregulated by ShvR and CciRd | cciR− shvR− | 4 |
| cciR− shvR+ | 135 | |
| cciR+ shvR− | 14 | |
| cciR+ shvR+ | 3 |
Total number of genes positively or negatively regulated in stationary-phase cultures of K56-2shvR::Tp that are regulated differently from genes in K56-2.
Number of genes positively or negatively regulated in stationary-phase cultures of K56-2shvR::Tp that are regulated differently from genes in K56-2 but unaffected in K56-R2 (cepR) and K56-2cciR (41).
Number of genes positively or negatively coregulated in stationary-phase cultures of K56-R2 (cepR) and K56-2shvR::Tp mutants.
Number of genes positively or negatively coregulated in stationary-phase cultures of K56-2cciR and K56-2shvR::Tp mutants.
Numbers of coregulated genes were grouped as follows: +, positive regulation; −, negative regulation. Previously published data from O'Grady et al., 2009 (41), regarding number of genes positively or negatively regulated in stationary-phase cultures of K56-R2 (cepR) or K56-2cciR that were regulated differently in K56-2 were used for comparison with K56-2shvR::Tp.
TABLE 4.
Microarray and qRT-PCR analysis of selected genes showing differential expression in K56-2shvR::Tp and K56-2
| Gene | Functiona | Fold changeb |
|
|---|---|---|---|
| Microarray | qRT-PCR | ||
| BCAM1870 | AHL synthase CepI | NC | 3.6 |
| BCAM1868 | AHL regulator CepR | NC | 5.6 |
| BCAM0239a | AHL synthase CciI | 1.8 | 12.5c |
| BCAM0240 | AHL regulator CciR | NC | 12.5c |
| BCAS0204 | ABC transporter ATP-binding protein | −100d | −34.5 |
| BCAS0215 | Putative exported protein | −100 | −51.7 |
| BCAS0220 | Putative permease | −38.6 | −193.4 |
| BCAM2307 | Zinc metalloprotease ZmpB | −4.4 | ND |
| BCAS0196 | Putative polygalacturonase | 2.3 | ND |
Function derived from B. cenocepacia J2315 (21).
Change in stationary-phase cultures of K56-2shvR::Tp compared to those of K56-2 as determined by microarray analysis or using qRT-PCR. NC, no change; ND, not determined using qRT-PCR.
cciI and cciR are cotranscribed (36).
Change in expression of BCAS0204 was included, as it is predicted to be part of the afcA operon (60), although it did not meet statistical significance for microarray analysis (detected as “absent” in K56-2shvR::Tp).
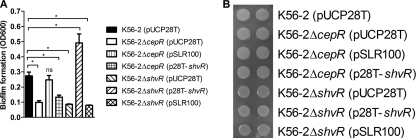
Both cepR and shvR mutants have reduced biofilm formation (4, 54) compared to that of the wild type. Since many genes were coregulated by both CepR and ShvR, we sought to determine whether cross-complementation with the other regulator on a high-copy plasmid would allow restoration of biofilm formation or colony morphology changes in cepR or shvR mutants. Reduced biofilm formation of the cepR mutant was restored by expression of cepR, but not shvR, in trans. Similarly, reduced biofilm formation of the shvR mutant was restored by expression of shvR, but not cepR, in trans (Fig. 3 A). These data suggest that CepR and ShvR independently regulate genes contributing to biofilm formation, as neither regulator can compensate the other for the biofilm defect. Colony morphology was assessed in the cepR and shvR mutants following growth on agar for 72 h at 28°C (data not shown) and 37°C. The shvR mutant exhibited shiny colony morphology, while the cepR mutant did not differ from K56-2 and appeared rough (Fig. 3B). Expression of cepR or shvR in trans in the cepR mutant had no effect on colony morphology, as cells remained rough. Rough colony morphology was restored in the shvR mutant by expression of shvR, but not cepR, in trans (Fig. 3B). This suggests that ShvR is required for rough colony morphology, and a defect in shvR cannot be compensated by overexpression of cepR.
FIG. 3.
Regulation of biofilm formation and colony morphology. (A) Biofilm formation was assessed on polystyrene pegs. Statistical significance was determined (*, P < 0.001 [one-way ANOVA]). All values are the means ± SD of results from 8 replicate cultures and are representative of results from at least two individual trials. (B) Colony morphology of cultures spot inoculated in 8 replicates onto LB agar. Images are representative of results from two individual trials.
ShvR negatively regulates protease and lipase activities and the type II secretion system (T2SS).
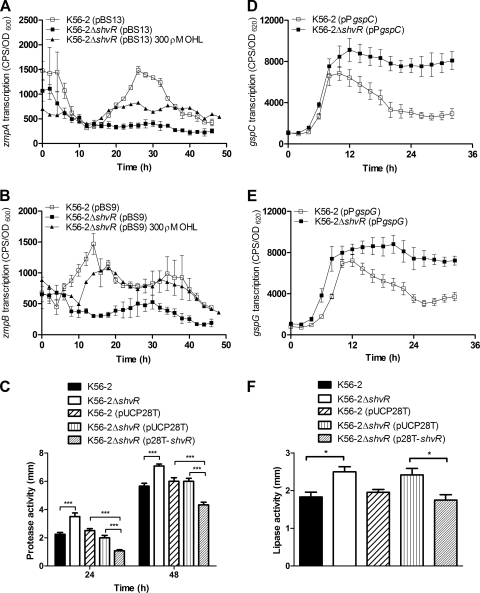
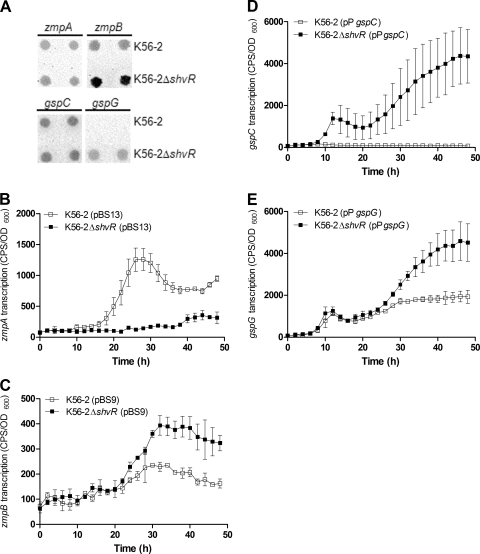
Analysis of zmpA and zmpB mutants in a rat chronic respiratory infection model showed that ZmpA and ZmpB contribute significantly to virulence and suggests that ZmpA positively influences persistence (10, 26). Microarray analysis showed that zmpB expression was significantly decreased in the shvR mutant compared to that in K56-2 (Table 4). Expression of both zmpA and zmpB promoter::lux fusions was significantly decreased in the shvR mutant compared to that in K56-2 in LB (Fig. 4 A and B) and PTSB (data not shown).
FIG. 4.
Expression of zmpA, zmpB, gspC, and the gspG operon and protease and lipase activities. Expression of zmpA (A), zmpB (B), gspC (D), or gspG (E) was monitored using promoter::lux fusions in B. cenocepacia in LB at 37°C. Expression is significantly decreased in K56-2ΔshvR compared to that in K56-2 from 20 to 37.5 h for zmpA and from 8 to 21.5 h for zmpB. Expression is significantly increased in K56-2ΔshvR compared to that in K56-2 from 16 to 32 h for gspC and gspG. Expression is significantly increased in K56-2ΔshvR including 300 pM OHL compared to that in K56-2ΔshvR from 17.5 to 26.5 h and 30.5 to 48 h for zmpA and 12 to 23.5 h and 32 to 40.5 h for zmpB (P < 0.001, two-way ANOVA). All values are the means ± SD of results from triplicate cultures and are representative of results from at least two individual trials. Cultures were spot inoculated in triplicate on D-BHI-1.5% skim agar for protease activity (C) or 1% Tween 80 agar for lipase activity (F), and zones of clearing or precipitation around colony growth were measured, respectively. Statistical significance was determined (*, P < 0.05; ***, P < 0.001 [one-way ANOVA]).
Regulation of zmpA and zmpB by ShvR was further investigated using phenotypic assays. In contrast to the gene expression data, protease activity was significantly increased in the shvR mutant compared to that in K56-2 on D-BHI-1.5% skim milk agar after a 24- and 48-h incubation (Fig. 4C). When shvR was expressed on the multicopy pUCP28T plasmid, protease activity was significantly decreased in K56-2ΔshvR(p28T-shvR) compared to that in either K56-2(pUCP28T) or K56-2ΔshvR(pUCP28T) (Fig. 4C). Data from these experiments suggest that ShvR negatively regulates protease activity. ZmpA and ZmpB are secreted via the T2SS, which is comprised of at least 12 open reading frames (ORFs), including gspC, and the gspD and gspG operons. In order to reconcile the apparent contradiction between transcriptional and phenotypic data, we analyzed expression of the T2SS genes. The expression of gspC and the gspG promoter::lux fusions was significantly increased in the shvR mutant compared to that in K56-2 (Fig. 4D and E). To determine whether increased expression of the T2SS genes in the shvR mutant influenced activity of additional proteins secreted by the T2SS, lipase assays were performed. Lipase activity was significantly increased in the shvR mutant compared to that in K56-2 when using 1% Tween 80 (Fig. 4F) or 1% tributyrin (data not shown) as substrates. When shvR was expressed in trans, lipase activity was significantly decreased in K56-2ΔshvR(p28T-shvR) compared to that in K56-2ΔshvR(pUCP28T) (Fig. 4F). Additionally, the expression of putative polygalacturonase encoded by BCAS0196 was significantly increased in the shvR mutant compared to that in the wild type (Table 4). Therefore, expression of at least four enzymes secreted by the T2SS was increased in the shvR mutant compared to that in the wild type, suggesting that the increased protease and lipase activity could be primarily due to ShvR regulation of the T2SS.
To further investigate the effects of ShvR on the expression of extracellular zinc metalloprotease and T2SS genes, strains carrying promoter::lux fusions were visualized for lux activity directly on skim milk agar plates. The intensities of the zmpA and gspC promoter::lux fusion activity appeared equivalent in K56-2 and the shvR mutant; however, in support of the protease activity data (Fig. 4C), and in contrast to results obtained in LB liquid cultures (Fig. 4B), zmpB promoter::lux fusion activity was increased in the shvR mutant compared to that in K56-2 (Fig. 5 A). Increased expression of gspG was also detected in the shvR mutant compared to that in K56-2 (Fig. 5A). Expression of zmpA, zmpB, gspC, and gspG was more easily detected using promoter::lux fusions on D-BHI-1.5% skim milk agar than using L agar plates, suggesting medium-dependent effects (data not shown). To address the regulation by ShvR in different media, promoter::lux fusions were compared in the medium constituents used to make D-BHI-1.5% skim milk. Expression of zmpA was significantly decreased in the shvR mutant compared to that in K56-2 in D-BHI-1.5% skim milk (Fig. 5B) or D-BHI or 1.5% skim milk (data not shown). In contrast, expression of zmpB, gspC, and gspG was significantly increased in the mutant compared to that in the wild type in D-BHI-1.5% skim milk (Fig. 5C to E) or D-BHI or 1.5% skim milk (data not shown). ZmpB has previously been shown to be more potent than ZmpA at degrading casein in the D-BHI-1.5% skim milk agar assay (26). Increased expression of zmpB rather than zmpA, in addition to significantly increased expression of the T2SS, most likely explains the increase in protease activity. Since CepR positively regulates zmpA and zmpB expression, we also considered the possibility that premature AHL activity in the shvR mutant led to deregulated control of zmpA and zmpB expression by CepR. The exogenous addition of 300 ρM OHL immediately prior to maximal cepR transcription in log phase cultures of the shvR mutant led to significantly increased zmpA and zmpB expression (Fig. 4A and B). This trend was not observed when 3,000 pM OHL was added to log-phase cultures or 300 pM OHL was added to stationary-phase (18-h) cultures of the shvR mutant (data not shown). These data suggest that appropriate timing and concentrations of exogenous OHL supplementation influenced zmpA and zmpB expression in the shvR mutant.
FIG. 5.
Expression of zmpA, zmpB, gspC, and the gspG operon on D-BHI-1.5% skim milk agar and in D-BHI-1.5% skim milk. (A) Expression of zmpA, zmpB, gspC, and gspG was monitored using promoter::lux fusions in B. cenocepacia from cultures spot inoculated onto D-BHI-1.5% skim milk agar. Images are representative of results from two individual trials. Expression of zmpA (B), zmpB (C), gspC (D), or gspG (E) was monitored using promoter::lux fusions in B. cenocepacia in D-BHI-1.5% skim milk at 37°C. Expression is significantly decreased in K56-2ΔshvR compared to that in K56-2 from 18 to 48 h for zmpA. Expression is significantly increased in K56-2ΔshvR compared to that in K56-2 from 29.5 to 48 h for zmpB, 38.5 to 48 h for gspC, and 35.5 to 48 h for gspG (P < 0.001, two-way ANOVA). All values are the means ± SD of results from triplicate cultures and are representative of results from at least two individual trials.
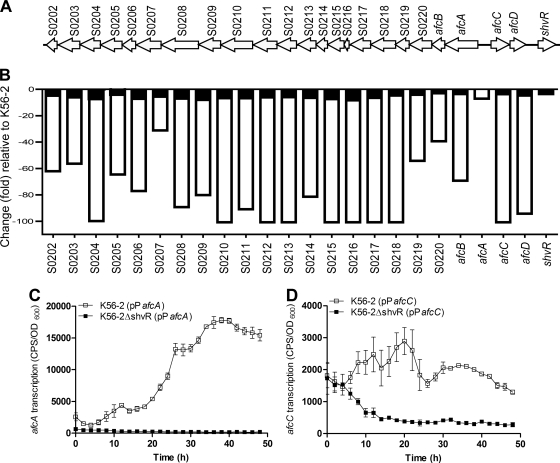
Expression of the afcA and afcC operons is ShvR dependent, and ShvR is required for antifungal activity.
We recently reported that CepR and CciR reciprocally regulate genes in the 24-kb afc genomic region adjacent to shvR (41). All 23 genes located in the afc genomic region had reduced expression in the shvR mutant compared to that in K56-2 according to microarray analysis (Fig. 6 A and B). Gene expression levels were reduced up to 100-fold, indicating that these were the most highly regulated genes in the shvR mutant. Bioinformatic analysis predicted that the afcA operon contains 21 genes, whereas the divergently transcribed afcC operon contains two genes (60). Three genes located in different parts of the afcA operon were selected for expression studies. Reduced expression of these genes was confirmed in the shvR mutant by using qRT-PCR (Table 4). Furthermore, analysis of afcA and afcC promoter::lux fusions confirmed significantly lower expression of these operons in the shvR mutant than in K56-2 (Fig. 6C and D).
FIG. 6.
Genomic organization and expression of genes in the shvR/afc genomic region. (A) ShvR lies in the genomic region adjacent to the afcA and afcC operons, which are transcribed divergently. (B) Expression was measured in K56-2shvR::Tp (open bars) and K56-R2 (closed bars) compared to that of K56-2 as determined by microarray analysis. Changes in expression of BCAS0204 and BCAS0205 were included, as they are predicted to be part of the afcA operon (60), although they did not meet statistical significance for microarray analysis. Expression of afcA (C) and afcC (D) was monitored using promoter::lux fusions in B. cenocepacia in LB at 37°C. Expression is significantly decreased in K56-2ΔshvR versus K56-2 (P < 0.001, two-way ANOVA). All values are the means ± SD of results from triplicate cultures and are representative of results from at least two individual trials.
Mean expression levels of genes in the afcA and afcC operons were reduced 5.7-fold in the cepR mutant (41) compared to 77.6-fold in the shvR mutant (Fig. 6B). CepR positively regulates shvR (41); thus, we considered the possibility that CepR may indirectly regulate the afcA operon via positive regulation of ShvR. Cross-complementation experiments were performed to investigate expression of the afcA operon in the cepR or shvR mutant backgrounds with cepR or shvR provided in trans. Three genes located in different parts of the afcA operon were selected for expression studies. In the cepR mutant background, expression of these genes was reduced between 3.5- and 15.3-fold (Table 5). Expression of these genes increased to levels close to or above parental levels by the presence of either cepR or shvR in trans (Table 5). In the shvR mutant background, expression of these genes was reduced between 106.5- and 242.4-fold and was restored to above parental levels by expression of shvR in trans (Table 5). In contrast, expression of cepR in trans in the shvR mutant background had little effect, in that gene expression levels remained between 69- and 211-fold reduced compared to those of the control (Table 5).
TABLE 5.
Expression of selected genes in the afcA operon
| Gene | Fold changea |
|||||
|---|---|---|---|---|---|---|
| K56- 2ΔcepR(pUCP28T) | K56- 2ΔcepR(pSLR100) | K56- 2ΔcepR(p28T-shvR) | K56- 2ΔshvR(pUCP28T) | K56- 2ΔshvR(p28T-shvR) | K56- 2ΔshvR(pSLR100) | |
| S0220 | −4.4 | 9.3 | −1.2 | −242.4 | 3.0 | −210.6 |
| S0208 | −15.3 | 4.9 | −5.2 | −214.1 | 2.9 | −100.9 |
| S0204 | −3.5 | 16.4 | −1.3 | −106.5 | 1.4 | −69.3 |
Change (fold) in stationary-phase cultures of cepR and shvR mutants carrying pUCP28T derivatives as indicated compared to cultures of K56-2(pUCP28T) as determined by qRT-PCR.
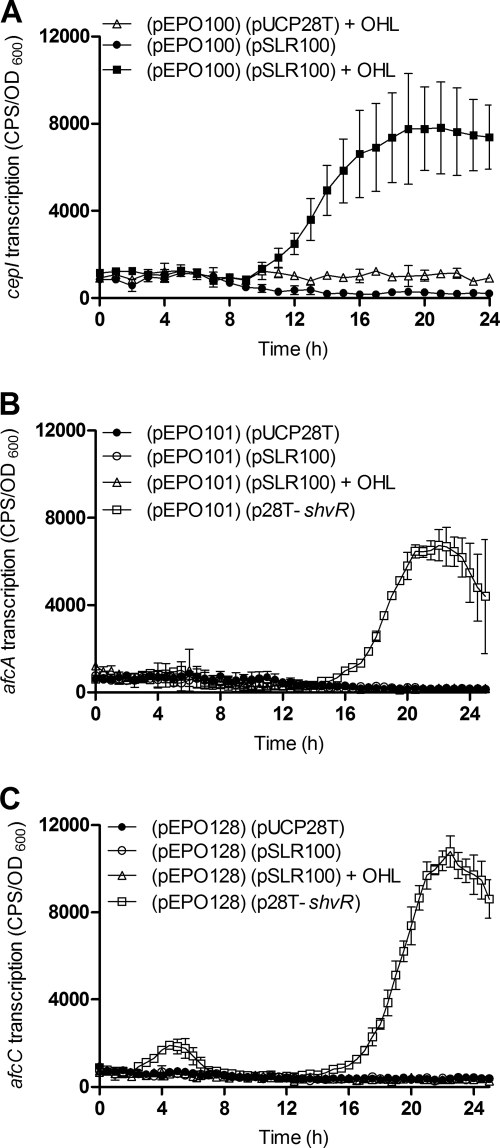
A heterologous host E. coli expression reporter system was developed to examine the potential for direct regulation of afcA and afcC by CepR and ShvR. E. coli strains were generated containing two types of plasmids: the first plasmid for expression of CepR or ShvR from pUCP28T in parallel with a second plasmid containing a promoter::lux fusion to enable measurement of promoter activity for cepI, afcA, afcC, or shvR. CepR binds the cepI promoter in the presence of OHL (59) and thus acted as a positive control in this E. coli expression reporter system. As expected, CepR activated expression of the cepI promoter::lux fusion only in the presence of OHL (Fig. 7 A) in a concentration-dependent manner (data not shown). CepR in the presence of OHL did not activate expression of promoter::lux fusions to afcA, afcC (Fig. 7B and C), or shvR in E. coli (data not shown), suggesting that CepR does not directly activate these promoters. In contrast, ShvR activated expression of afcA and afcC promoter::lux fusions (Fig. 7B and C). These data provide evidence that ShvR directly regulates the afcA and afcC promoter regions and suggest that CepR indirectly positively regulates these genes via positive regulation of ShvR.
FIG. 7.
Expression of cepI, afcA, and afcC in the heterologous host E. coli by using an expression-reporter system. Strains harbor two plasmids: a derivative of pUCP28T and a promoter::lux fusion for cepI (pEPO100), afcA (pEPO101), or afcC (pEPO128) as indicated. Assays were performed in the absence or presence of 3,000 pM OHL at 37°C. Expression of cepI (A), afcA (B), or afcC (C) was monitored using the appropriate promoter::lux fusion in E. coli also carrying pUCP28T, or cepR in trans (pSLR100) or shvR in trans (p28T-shvR). All values are the means ± SD of results from triplicate cultures and are representative of results from at least two individual trials.
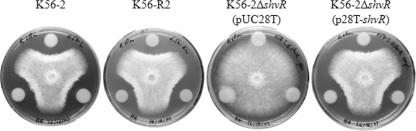
Mutations affecting afcA, afcC, and afcD in B. pyrrocinia BC11 were previously shown to impair production of an antifungal compound and inhibit antifungal activity against the soilborne phytopathogen Rhizoctonia solani (25). B. pyrrocinia BC11 AfcA, AfcC, and AfcD show greater than 95% identity with corresponding proteins in B. cenocepacia J2315, AU1054, HI2424, and MCO-3 (60). Considering the regulation by CepR and ShvR of genes in the afc genomic region, we compared antifungal activity of cepR and shvR mutants to that in the wild type. Antifungal activity against R. solani was detectable to a similar extent in K56-2 and the cepR mutant (Fig. 8). K56-2ΔshvR(pUCP28T) did not appear to have any antifungal activity; however, activity was restored to parental levels in K56-2ΔshvR(p28T-shvR) (Fig. 7). In summary, these data suggest that ShvR-dependent expression of the afcA and afcC operons was essential for the antifungal activity of K56-2.
FIG. 8.
Antifungal activity against R. solani. R. solani was grown on malt agar in the presence of K56-2, K56-R2 (cepR), K56-2ΔshvR(pUCP28T), or K56-2ΔshvR(p28T-shvR). Fungal growth inhibition was recorded after 3 days. The assay was performed in triplicate; a representative plate for each assay is shown.
DISCUSSION
We previously reported an alteration of virulence-related phenotypes in B. cenocepacia that was associated with a mutation of the ORF encoding the LTTR ShvR (4). In this study, we characterized ShvR regulation and determined that it exerts negative autoregulation, a feature typical of LTTRs. ShvR also exerts the transcriptional control of genes that are located in the genomic region adjacent to shvR and genes located on all three chromosomes and the plasmid, as well as QS-regulated genes. The cepIR and cciIR genes are negatively regulated by ShvR, and consequently ShvR has a significant influence on the timing of expression of AHL activity in that AHLs accumulate earlier in an shvR mutant than in the wild type, although total AHL activity is similar between the strains. The reduced AHL activity in the shvR mutant during stationary phase likely contributed to the identification of numerous genes coregulated by CepR and ShvR, including zmpA, zmpB, and the afcA and afcC operons. Despite the identification of coregulated genes, biofilm formation and colony morphology are independently regulated by CepR and ShvR. More than 67% of the total number of genes that were differentially expressed in the shvR mutant were not altered in the cepR or cciR mutants. Independent ShvR-mediated regulation of these genes may play a role in the altered biofilm formation and colony morphology observed in the shvR mutant. Reduced alfalfa virulence by shvR but not cepI mutants of B. cenocepacia K56-2 provides additional evidence for independent regulation by ShvR (4, 55).
ZmpA and ZmpB extracellular proteases each contribute to virulence in a rat chronic respiratory infection model (26). Several studies highlight the importance of the T2SS, encoded by the gsp genes, for secretion of proteases and other extracellular enzymes (14, 17, 27, 52). Protease, phospholipase C, and hemolysin activities were reduced due to a gspN mutation in Burkholderia vietnamiensis CEP40 (14). B. cenocepacia H111 with mutations in the gspD or gspG operons failed to produce extracellular protease and had significantly reduced lipase activity (27). In B. cenocepacia AU1054, a gspJ mutation resulted in reduced protease and polygalacturonase activities (52). Analysis of sequenced genomes at http://www.burkholderia.com (60) indicates that B. cenocepacia J2315 is the only Burkholderia strain which lacks gspL, which is part of the gspG operon (21). B. cenocepacia J2315 produces little or no protease activity on skim milk agar despite detection of high zmpA expression levels using qRT-PCR (17).
QS regulation of the T2SS is observed in Pseudomonas aeruginosa and Burkholderia glumae. Expression of the XcpR protein in P. aeruginosa is most easily detected at the onset of stationary phase, after which protein levels decrease slightly (9). The B. glumae TofIR QS system positively regulates the IclR-type transcriptional regulator gene, QsmR, which directly binds the promoters for gspC, gspD, and gspG (18). We previously noted that expression of gspC and gspG was decreased in the wild type compared to that in a cepI mutant or upon addition of OHL to a cepI mutant, suggesting negative regulation by CepIR (53). The negative effect of CepIR on gspC and gspG is consistent with the decrease in gsp expression that occurs upon entry into stationary phase in wild-type cells. After a 24-h incubation, gspC and gspG operon transcription in the wild type is reduced to a steady-state level that remains higher than basal transcription levels observed in starting cultures. Expression of the P. aeruginosa xcp gene is activated by QS followed by a decrease in expression to a level that is higher than that of starting cultures (9). Together, these data suggest that expression of the T2SS is tightly regulated by QS in both B. cenocepacia and P. aeruginosa, and once the apparatus is fully formed, gene expression is maintained to facilitate continued secretion by the T2SS. Expression of T2SS genes was significantly increased in the shvR mutant compared to that in the wild type during stationary phase of growth, when AHL activity was reduced. It is possible that unidentified ShvR-regulated factors, in addition to altered AHL activity in the shvR mutant, contribute to the sustained and increased expression of the T2SS that resulted in increased protease and lipase activities.
Reduced zmpA and zmpB promoter activity in the shvR mutant in LB cultures was recovered by supplementation of exogenous OHL immediately prior to maximal cepR transcription. These data are in concordance with a positive regulatory effect of CepR and provide evidence that reduced expression of these genes in the shvR mutant in LB occurred via ShvR influence on AHL activity and QS gene expression. The absence of shvR had a different effect on zmpB expression in different media. The combination of D-BHI-1.5% skim milk and the shvR mutation resulted in increased zmpB expression. These conditions may provide a stimulus for an intermediate regulator of zmpB that functions in the absence of ShvR, resulting in increased zmpB expression. Previous studies demonstrated that CciR regulation of zmpA and zmpB is influenced by medium and growth phase, indicating that the regulation of these extracellular enzymes is complex (26, 36, 41). B. cenocepacia produces cis-2-dodecenoic acid (Burkholderia diffusible signal factor [BDSF]), which is a QS signal (7, 11, 38, 43). While zmpB expression was not examined, BDSF positively regulated zmpA and lipAB (lipase) expression in wild-type and cepR mutant backgrounds (11). These data indicate that the BDSF signal is another factor that regulates expression of these extracellular enzymes.
We previously showed that the shvR/afc genomic region is positively regulated by CepR and negatively regulated by CciR (41). During review of the manuscript, expression of shvR and the afcA and afcC operons was reported to be decreased 2.6-fold in a B. cenocepacia J2315 BDSF synthase mutant (BCAM0581 [rpfF]) (38). We previously reported a 5.7-fold decrease in expression of these genes in a K56-2 cepR mutant (41). In contrast to the effects of QS, expression of the afcA and afcC operons was substantially lower in the shvR mutant (77.6-fold) than in the wild type. Data from cross-complementation experiments and the heterologous host expression reporter system suggested that the expression of the afcA and afcC operons is ShvR dependent and that the effects of the CepIR system on these operons are likely indirectly controlled via ShvR.
The majority of genes in the afcA operon are uncharacterized, although in B. pyrrocinia BC11, afcA and afcCD were shown to be involved in the production of an antifungal with inhibitory activity against the soilborne phytopathogens Rhizoctonia solani and Pythium ultimum (25). Antifungal activity was substantially reduced in the shvR mutant, likely due to the reduced expression of genes in the afc genomic region. Antifungal production is QS dependent in certain strains of Burkholderia ambifaria, Burkholderia pyrrocinia, and Burkholderia lata (46) but was not altered by the mutation of cepR in B. cenocepacia K56-2, providing evidence of species variation in antifungal activity. It is noteworthy that the B. pyrrocinia BC11 antifungal activity was not found against pathogenic fungi more commonly associated with human infections such as Candida albicans or Aspergillus species (25). We determined that shvR expression was highest after extended incubation at 29°C, which is consistent with the observation that production of antifungal activity in B. pyrrocinia was detected at 30°C (25). Genes in the shvR/afc genomic region are more highly expressed in the agricultural field isolate B. cenocepacia HI2424 in soil-like culture conditions at 22°C than in synthetic CF sputum medium at 37°C (61) and in the CF isolate B. cenocepacia J2315 in soil-like culture conditions than in synthetic CF sputum medium at 37°C (62). Together, these data suggest that in addition to ShvR contribution to virulence in plant and animal infection models, ShvR control of the antifungal encoded by afc genes may play an important role in the competitive environment of the plant rhizosphere, protecting plants against phytopathogenic fungi.
ShvR exhibited features typical of LTTRs, namely, negative autoregulation and regulation of genes located in the adjacent afc genomic region. Negative autoregulation may explain the biphasic expression pattern of shvR observed in cultures grown at 29°C. The CepIR and CciIR QS systems display a complex regulatory interrelationship that involves positive or negative autoregulation, CepR-dependent expression of cciIR, and negative regulation of cepI by CciR (29, 36). While shvR expression is inversely regulated by CepIR and CciIR (41), a feedback loop exists where ShvR negatively regulates cepIR and cciIR expression. A situation may exist where basal shvR expression at low cell densities is sufficient to repress cepIR and cciIR expression and delay AHL accumulation until positive regulators of cepIR and cciIR become active as cell density increases. Apart from the AHL-dependent effects of ShvR, global transcriptional changes observed in the shvR mutant may stem from ShvR regulation of intermediate regulators, including 18 LTTRs identified by using microarray analysis. Characterization of ShvR cofactor(s) may enable the identification of subsets of genes regulated under specific environmental conditions. In the current study, two promoters were determined to be directly regulated by ShvR. The identification of an ShvR-binding motif may reveal other promoters that are directly regulated versus those that are indirectly regulated. Further studies are under way to determine the mechanisms by which ShvR regulates virulence traits in B. cenocepacia.
Acknowledgments
These studies were supported by research grants from the Canadian Cystic Fibrosis Foundation (CCFF), Cystic Fibrosis Foundation Therapeutics (CFFT) (grant SOKOL06V0), and Canadian Institutes of Health Research (grant MOP-42510) to P.A.S. E.P.O. was the recipient of a CCFF fellowship.
Microarray processing was provided by the Mahenthiralingam Laboratory, Cardiff University, Wales, and initial data analysis by the Center for Bioinformatics of University of North Carolina at Chapel Hill, North Carolina, with support from CFFT.
Footnotes
Published ahead of print on 22 October 2010.
REFERENCES
- 1.Ausubel, F., R. Brent, R. Kingston, D. Moore, J. Seidman, J. Smith, and K. Struhl. 1989. Current protocols in molecular biology, vol. 1. John Wiley & Sons, Inc., New York, NY.
- 2.Baldwin, A., P. A. Sokol, J. Parkhill, and E. Mahenthiralingam. 2004. The Burkholderia cepacia epidemic strain marker is part of a novel genomic island encoding both virulence and metabolism-associated genes in Burkholderia cenocepacia. Infect. Immun. 72:1537-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernier, S. P., A. L. Beeston, and P. A. Sokol. 2008. Detection of N-acyl homoserine lactones using a traI-luxCDABE-based biosensor as a high-throughput screening tool. BMC Biotechnol. 8:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernier, S. P., D. T. Nguyen, and P. A. Sokol. 2008. A LysR-type transcriptional regulator in Burkholderia cenocepacia influences colony morphology and virulence. Infect. Immun. 76:38-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bernier, S. P., and P. A. Sokol. 2005. Use of suppression-subtractive hybridization to identify genes in the Burkholderia cepacia complex that are unique to Burkholderia cenocepacia. J. Bacteriol. 187:5278-5291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bjarnason, J., C. M. Southward, and M. G. Surette. 2003. Genomic profiling of iron-responsive genes in Salmonella enterica serovar Typhimurium by high-throughput screening of a random promoter library. J. Bacteriol. 185:4973-4982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boon, C., Y. Deng, L. H. Wang, Y. He, J. L. Xu, Y. Fan, S. Q. Pan, and L. H. Zhang. 2008. A novel DSF-like signal from Burkholderia cenocepacia interferes with Candida albicans morphological transition. ISME J. 2:27-36. [DOI] [PubMed] [Google Scholar]
- 8.Chambers, C. E., E. I. Lutter, M. B. Visser, P. P. Law, and P. A. Sokol. 2006. Identification of potential CepR regulated genes using a cep box motif-based search of the Burkholderia cenocepacia genome. BMC Microbiol. 6:104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chapon-Herve, V., M. Akrim, A. Latifi, P. Williams, A. Lazdunski, and M. Bally. 1997. Regulation of the xcp secretion pathway by multiple quorum-sensing modulons in Pseudomonas aeruginosa. Mol. Microbiol. 24:1169-1178. [DOI] [PubMed] [Google Scholar]
- 10.Corbett, C. R., M. N. Burtnick, C. Kooi, D. E. Woods, and P. A. Sokol. 2003. An extracellular zinc metalloprotease gene of Burkholderia cepacia. Microbiology 149:2263-2271. [DOI] [PubMed] [Google Scholar]
- 11.Deng, Y., C. Boon, L. Eberl, and L. H. Zhang. 2009. Differential modulation of Burkholderia cenocepacia virulence and energy metabolism by the quorum-sensing signal BDSF and its synthase. J. Bacteriol. 191:7270-7278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dennis, J. J., and P. A. Sokol. 1995. Electrotransformation of Pseudomonas. Methods Mol. Biol. 47:125-133. [DOI] [PubMed] [Google Scholar]
- 13.Duan, K., C. Dammel, J. Stein, H. Rabin, and M. G. Surette. 2003. Modulation of Pseudomonas aeruginosa gene expression by host microflora through interspecies communication. Mol. Microbiol. 50:1477-1491. [DOI] [PubMed] [Google Scholar]
- 14.Fehlner-Gardiner, C. C., T. M. Hopkins, and M. A. Valvano. 2002. Identification of a general secretory pathway in a human isolate of Burkholderia vietnamiensis (formerly B. cepacia complex genomovar V) that is required for the secretion of hemolysin and phospholipase C activities. Microb. Pathog. 32:249-254. [DOI] [PubMed] [Google Scholar]
- 15.Figurski, D. H., and D. R. Helinski. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. U. S. A. 76:1648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flannagan, R. S., T. Linn, and M. A. Valvano. 2008. A system for the construction of targeted unmarked gene deletions in the genus Burkholderia. Environ. Microbiol. 10:1652-1660. [DOI] [PubMed] [Google Scholar]
- 17.Gingues, S., C. Kooi, M. B. Visser, B. Subsin, and P. A. Sokol. 2005. Distribution and expression of the ZmpA metalloprotease in the Burkholderia cepacia complex. J. Bacteriol. 187:8247-8255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goo, E., Y. Kang, H. Kim, and I. Hwang. 2010. Proteomic analysis of quorum sensing-dependent proteins in Burkholderia glumae. J. Proteome Res. 9:3184-3199. [DOI] [PubMed] [Google Scholar]
- 19.Gotschlich, A., B. Huber, O. Geisenberger, A. Togl, A. Steidle, K. Riedel, P. Hill, B. Tummler, P. Vandamme, B. Middleton, M. Camara, P. Williams, A. Hardman, and L. Eberl. 2001. Synthesis of multiple N-acylhomoserine lactones is widespread among the members of the Burkholderia cepacia complex. Syst. Appl. Microbiol. 24:1-14. [DOI] [PubMed] [Google Scholar]
- 20.Hoang, T. T., R. R. Karkhoff-Schweizer, A. J. Kutchma, and H. P. Schweizer. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77-86. [DOI] [PubMed] [Google Scholar]
- 21.Holden, M. T., H. M. Seth-Smith, L. C. Crossman, M. Sebaihia, S. D. Bentley, A. M. Cerdeno-Tarraga, N. R. Thomson, N. Bason, M. A. Quail, S. Sharp, I. Cherevach, C. Churcher, I. Goodhead, H. Hauser, N. Holroyd, K. Mungall, P. Scott, D. Walker, B. White, H. Rose, P. Iversen, D. Mil-Homens, E. P. Rocha, A. M. Fialho, A. Baldwin, C. Dowson, B. G. Barrell, J. R. Govan, P. Vandamme, C. A. Hart, E. Mahenthiralingam, and J. Parkhill. 2009. The genome of Burkholderia cenocepacia J2315, an epidemic pathogen of cystic fibrosis patients. J. Bacteriol. 191:261-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huber, B., F. Feldmann, M. Kothe, P. Vandamme, J. Wopperer, K. Riedel, and L. Eberl. 2004. Identification of a novel virulence factor in Burkholderia cenocepacia H111 required for efficient slow killing of Caenorhabditis elegans. Infect. Immun. 72:7220-7230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huber, B., K. Riedel, M. Hentzer, A. Heydorn, A. Gotschlich, M. Givskov, S. Molin, and L. Eberl. 2001. The cep quorum-sensing system of Burkholderia cepacia H111 controls biofilm formation and swarming motility. Microbiology 147:2517-2528. [DOI] [PubMed] [Google Scholar]
- 24.Isles, A., I. Maclusky, M. Corey, R. Gold, C. Prober, P. Fleming, and H. Levison. 1984. Pseudomonas cepacia infection in cystic fibrosis: an emerging problem. J. Pediatr. 104:206-210. [DOI] [PubMed] [Google Scholar]
- 25.Kang, Y., R. Carlson, W. Tharpe, and M. A. Schell. 1998. Characterization of genes involved in biosynthesis of a novel antibiotic from Burkholderia cepacia BC11 and their role in biological control of Rhizoctonia solani. Appl. Environ. Microbiol. 64:3939-3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kooi, C., B. Subsin, R. Chen, B. Pohorelic, and P. A. Sokol. 2006. Burkholderia cenocepacia ZmpB is a broad-specificity zinc metalloprotease involved in virulence. Infect. Immun. 74:4083-4093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kothe, M., M. Antl, B. Huber, K. Stoecker, D. Ebrecht, I. Steinmetz, and L. Eberl. 2003. Killing of Caenorhabditis elegans by Burkholderia cepacia is controlled by the cep quorum-sensing system. Cell. Microbiol. 5:343-351. [DOI] [PubMed] [Google Scholar]
- 28.Leiske, D. L., A. Karimpour-Fard, P. S. Hume, B. D. Fairbanks, and R. T. Gill. 2006. A comparison of alternative 60-mer probe designs in an in situ-synthesized oligonucleotide microarray. BMC Genomics 7:72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lewenza, S., B. Conway, E. P. Greenberg, and P. A. Sokol. 1999. Quorum sensing in Burkholderia cepacia: identification of the LuxRI homologs CepRI. J. Bacteriol. 181:748-756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lewenza, S., and P. A. Sokol. 2001. Regulation of ornibactin biosynthesis and N-acyl-l-homoserine lactone production by CepR in Burkholderia cepacia. J. Bacteriol. 183:2212-2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Loutet, S. A., and M. A. Valvano. 2010. A decade of Burkholderia cenocepacia virulence determinant research. Infect. Immun. 78:4088-4100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lutter, E., S. Lewenza, J. J. Dennis, M. B. Visser, and P. A. Sokol. 2001. Distribution of quorum-sensing genes in the Burkholderia cepacia complex. Infect. Immun. 69:4661-4666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maddocks, S. E., and P. C. Oyston. 2008. Structure and function of the LysR-type transcriptional regulator (LTTR) family proteins. Microbiology 154:3609-3623. [DOI] [PubMed] [Google Scholar]
- 34.Mahenthiralingam, E., T. Coenye, J. W. Chung, D. P. Speert, J. R. Govan, P. Taylor, and P. Vandamme. 2000. Diagnostically and experimentally useful panel of strains from the Burkholderia cepacia complex. J. Clin. Microbiol. 38:910-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mahenthiralingam, E., T. A. Urban, and J. B. Goldberg. 2005. The multifarious, multireplicon Burkholderia cepacia complex. Nat. Rev. Microbiol. 3:144-156. [DOI] [PubMed] [Google Scholar]
- 36.Malott, R. J., A. Baldwin, E. Mahenthiralingam, and P. A. Sokol. 2005. Characterization of the cciIR quorum-sensing system in Burkholderia cenocepacia. Infect. Immun. 73:4982-4992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Malott, R. J., E. P. O'Grady, J. Toller, S. Inhulsen, L. Eberl, and P. A. Sokol. 2009. A Burkholderia cenocepacia orphan LuxR homolog is involved in quorum-sensing regulation. J. Bacteriol. 191:2447-2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McCarthy, Y., L. Yang, K. B. Twomey, A. Sass, T. Tolker-Nielsen, E. Mahenthiralingam, J. M. Dow, and R. P. Ryan. 2010. A sensor kinase recognizing the cell-cell signal BDSF (cis-2-dodecenoic acid) regulates virulence in Burkholderia cenocepacia. Mol. Microbiol. 77:1220-1236. [DOI] [PubMed] [Google Scholar]
- 39.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ng, W. L., and B. L. Bassler. 2009. Bacterial quorum-sensing network architectures. Annu. Rev. Genet. 43:197-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.O'Grady, E. P., D. F. Viteri, R. J. Malott, and P. A. Sokol. 2009. Reciprocal regulation by the CepIR and CciIR quorum sensing systems in Burkholderia cenocepacia. BMC Genomics 10:441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rozen, S., and H. Skaletsky. 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132:365-386. [DOI] [PubMed] [Google Scholar]
- 43.Ryan, R. P., Y. McCarthy, S. A. Watt, K. Niehaus, and J. M. Dow. 2009. Intraspecies signaling involving the diffusible signal factor BDSF (cis-2-dodecenoic acid) influences virulence in Burkholderia cenocepacia. J. Bacteriol. 191:5013-5019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saiman, L., and J. Siegel. 2004. Infection control in cystic fibrosis. Clin. Microbiol. Rev. 17:57-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 46.Schmidt, S., J. F. Blom, J. Pernthaler, G. Berg, A. Baldwin, E. Mahenthiralingam, and L. Eberl. 2009. Production of the antifungal compound pyrrolnitrin is quorum sensing-regulated in members of the Burkholderia cepacia complex. Environ. Microbiol. 11:1422-1437. [DOI] [PubMed] [Google Scholar]
- 47.Schweizer, H. P., and T. T. Hoang. 1995. An improved system for gene replacement and xylE fusion analysis in Pseudomonas aeruginosa. Gene 158:15-22. [DOI] [PubMed] [Google Scholar]
- 48.Schweizer, H. P., T. Klassen, and T. T. Hoang. 1996. Improved methods for gene analysis and expression in Pseudomonas spp., 1995/05/26 ed. ASM Press, Washington, DC.
- 49.Sokol, P. A., R. J. Malott, K. Riedel, and L. Eberl. 2007. Communication systems in the genus Burkholderia: global regulators and targets for novel antipathogenic drugs. Future Microbiol. 2:555-563. [DOI] [PubMed] [Google Scholar]
- 50.Sokol, P. A., D. E. Ohman, and B. H. Iglewski. 1979. A more sensitive plate assay for detection of protease production by Pseudomonas aeruginosa. J. Clin. Microbiol. 9:538-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sokol, P. A., U. Sajjan, M. B. Visser, S. Gingues, J. Forstner, and C. Kooi. 2003. The CepIR quorum-sensing system contributes to the virulence of Burkholderia cenocepacia respiratory infections. Microbiology 149:3649-3658. [DOI] [PubMed] [Google Scholar]
- 52.Somvanshi, V. S., P. Viswanathan, J. L. Jacobs, M. H. Mulks, G. W. Sundin, and T. A. Ciche. 2010. The type 2 secretion pseudopilin, gspJ, is required for multihost pathogenicity of Burkholderia cenocepacia AU1054. Infect. Immun. 78:4110-4121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Subsin, B., C. E. Chambers, M. B. Visser, and P. A. Sokol. 2007. Identification of genes regulated by the cepIR quorum-sensing system in Burkholderia cenocepacia by high-throughput screening of a random promoter library. J. Bacteriol. 189:968-979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tomlin, K. L., R. J. Malott, G. Ramage, D. G. Storey, P. A. Sokol, and H. Ceri. 2005. Quorum-sensing mutations affect attachment and stability of Burkholderia cenocepacia biofilms. Appl. Environ. Microbiol. 71:5208-5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Uehlinger, S., S. Schwager, S. P. Bernier, K. Riedel, D. T. Nguyen, P. A. Sokol, and L. Eberl. 2009. Identification of specific and universal virulence factors in Burkholderia cenocepacia strains by using multiple infection hosts. Infect. Immun. 77:4102-4110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vandamme, P., D. Henry, T. Coenye, S. Nzula, M. Vancanneyt, J. J. LiPuma, D. P. Speert, J. R. Govan, and E. Mahenthiralingam. 2002. Burkholderia anthina sp. nov. and Burkholderia pyrrocinia, two additional Burkholderia cepacia complex bacteria, may confound results of new molecular diagnostic tools. FEMS Immunol. Med. Microbiol. 33:143-149. [DOI] [PubMed] [Google Scholar]
- 57.Venturi, V., A. Friscina, I. Bertani, G. Devescovi, and C. Aguilar. 2004. Quorum sensing in the Burkholderia cepacia complex. Res. Microbiol. 155:238-244. [DOI] [PubMed] [Google Scholar]
- 58.Vergunst, A. C., A. H. Meijer, S. A. Renshaw, and D. O'Callaghan. 2010. Burkholderia cenocepacia creates an intramacrophage replication niche in zebrafish embryos, followed by bacterial dissemination and establishment of systemic infection. Infect. Immun. 78:1495-1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Weingart, C. L., C. E. White, S. Liu, Y. Chai, H. Cho, C. S. Tsai, Y. Wei, N. R. Delay, M. R. Gronquist, A. Eberhard, and S. C. Winans. 2005. Direct binding of the quorum sensing regulator CepR of Burkholderia cenocepacia to two target promoters in vitro. Mol. Microbiol. 57:452-467. [DOI] [PubMed] [Google Scholar]
- 60.Winsor, G. L., B. Khaira, T. Van Rossum, R. Lo, M. D. Whiteside, and F. S. Brinkman. 2008. The Burkholderia Genome Database: facilitating flexible queries and comparative analyses. Bioinformatics 24:2803-2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yoder-Himes, D. R., P. S. Chain, Y. Zhu, O. Wurtzel, E. M. Rubin, J. M. Tiedje, and R. Sorek. 2009. Mapping the Burkholderia cenocepacia niche response via high-throughput sequencing. Proc. Natl. Acad. Sci. U. S. A. 106:3976-3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yoder-Himes, D. R., K. T. Konstantinidis, and J. M. Tiedje. 2010. Identification of potential therapeutic targets for Burkholderia cenocepacia by comparative transcriptomics. PLoS One 5:e8724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhu, J., J. W. Beaber, M. I. More, C. Fuqua, A. Eberhard, and S. C. Winans. 1998. Analogs of the autoinducer 3-oxooctanoyl-homoserine lactone strongly inhibit activity of the TraR protein of Agrobacterium tumefaciens. J. Bacteriol. 180:5398-5405. [DOI] [PMC free article] [PubMed] [Google Scholar]