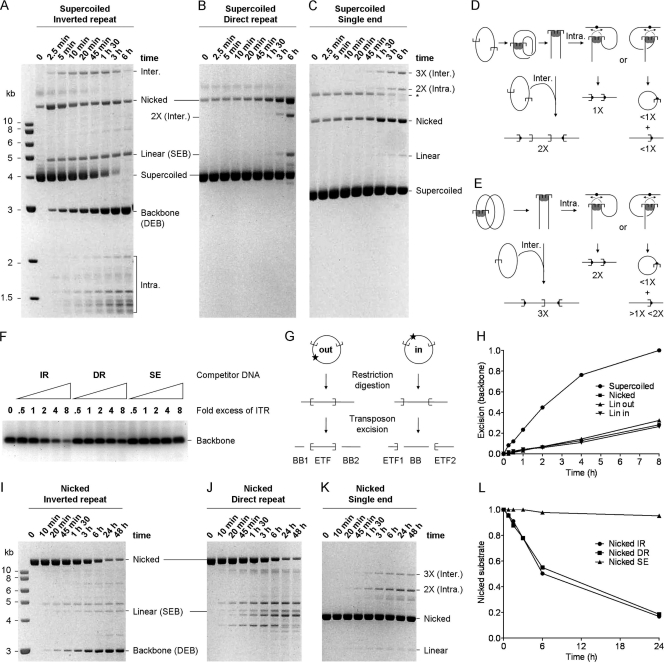
FIG. 2.
The configuration of transposon ends influences the kinetics of cleavage. (A) The kinetics of transposition with the standard inverted-repeat substrate (pRC650). Excision of the transposon yields the plasmid backbone, which is an end product of the reaction. A photograph of an ethidium bromide-stained agarose gel is shown. (B and C) Cleavage with the direct-repeat (pRC883) (B) and single-end (pRC919) (C) substrates are difficult to quantify, because there is no product equivalent to the backbone in the inverted-repeat reactions. (The products of the reactions corresponding to panels B and C are shown in panels D and E, respectively.) The only unique products are from the integration reaction. The direct repeat yields an intermolecular product twice the size of the substrate (2X). The single-end substrate product yields a 2X intramolecular integration product and a 3X intermolecular integration product. The simplest estimate of cleavage is given by the rate at which the substrate disappears. * indicates the supercoiled dimer form of the substrate. A photograph of an ethidium bromide-stained agarose gel is shown. (D) With a direct-repeat substrate, cleavage yields two linear fragments that together equal the size of the substrate. Intermolecular integration into an unreacted substrate plasmid, which is the only target available, generates a linear product two times the size of the substrate (2X). Intramolecular events produce two alternative outcomes depending on orientation of the target site: either they generate a linear product the size of the substrate (1X) or they generate a pair of products, namely, an open-circle product plus a linear product that together equal the size of the plasmid. (E) With a single-ended substrate, cleavage at the transposon end yields a linear intermediate of the same size as the substrate (1X). Intermolecular integration into an unreacted substrate plasmid, which is the only target available in these reactions, generates a linear product three times the size of the substrate (3X). Intramolecular events produce two alternative outcomes depending on the orientation of the target site: either they generate a linear product twice the size of the substrate (2X) or they generate a pair of products, namely, an open-circle product (<1X substrate) plus a corresponding linear product between one and two times the size of the substrate (>1X <2X). (F) A competition assay was performed using a principal substrate (pRC704) yielding an 800-bp backbone fragment, which can be quantified easily because it runs in a clear area of the gel. Reaction mixtures contained 2 nM principal substrate and 10 nM transposase. They were titrated up to an 8-fold molar excess of the competitor transposon ends. This is equivalent to an 8-fold molar excess of the double-ended plasmids and a 16-fold molar excess of the single-ended plasmid. All of the reactions were adjusted to contain the same total amount of DNA by the addition of pBluescript, where required. The only difference between any of the reactions was therefore the concentration of the respective competitor transposon ends. Cleavage of the principal substrate was analyzed on an agarose gel stained with SYBR green. IR, inverted repeat; DR, direct repeat; SE, single end. (G) The inverted-repeat plasmid was linearized with XmnI and NruI to yield the LinOUT and LinIN substrates, respectively. The excision products for these substrates are illustrated. Transposon excision from the LinOUT substrate yields two backbone (BB) fragments. (H) The kinetics of the excision reactions with the indicated substrates were analyzed by ethidium bromide-stained agarose gel electrophoresis and quantified using a Fluorimager (Fujifilm). “SC” and “Nicked” refer to the supercoiled and open circular forms of the inverted-repeat substrate used in panel A. LinOUT and LinIN are illustrated in panel G. Reactions were normalized to the level of backbone produced by the SC substrate and plotted. (I, J, and K) The kinetics of transposition reactions using nicked forms of the inverted-repeat (I), direct-repeat (J), and single-end (K) substrates. These substrates were prepared by treatment with a nicking endonuclease (Nb.BsrDI). For each time point, 100 ng DNA was loaded on the gel. The reactivity of the substrates can be estimated by assessing the disappearance of the nicked donor. (L) The nicked substrates described for panels I, J, and K were quantified as described for panel H. Values were normalized to the signal at time zero for each substrate.