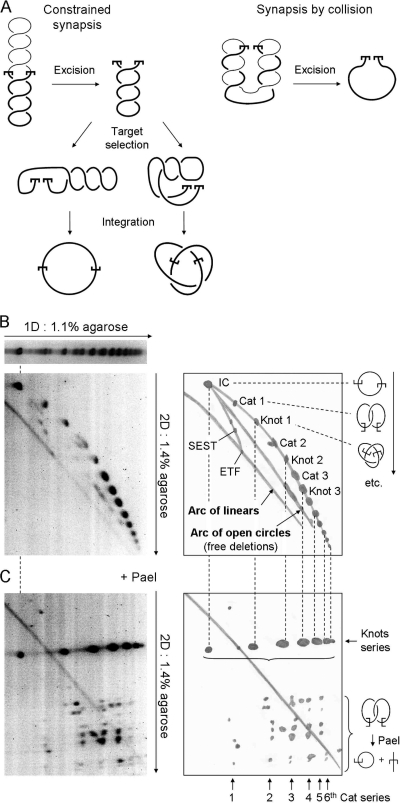
FIG. 4.
Synapsis in the plectosome. (A) Two models for the assembly of the mariner synaptic complex are illustrated. (Left panel) When synapsis of the transposon ends is constrained by the plectosome, the ETF retains the full superhelical density of the substrate. If intramolecular target sites are selected by random collision, any number of nodes may be trapped, ranging from zero up to the total number present. However, on average, half of the nodes are trapped in the product. (Right panel) If the transposon ends meet by random collision, unconstrained by the plectosome, any number of nodes may be trapped, up to the maximum number present. The illustration shows the situation when the transposon ends are at opposite ends of the plectosome when they collide: excision releases all of the nodes. However, we would expect collision to trap half of the nodes on average. Half of these would go on to be trapped in the product if the target site is acquired by collision. (B) The topology of intramolecular integration products obtained from a reaction with a negatively supercoiled inverted-repeat substrate (pRC1105) was analyzed by two-dimensional (2D) gel electrophoresis using the conditions indicated beside the gels. A drawing of each gel is provided to indicate the identities of the products. The substrate includes a 2.3-kb transposon and a 4.2-kb backbone. The gel conditions and the respective sizes of the transposon and backbone fragments are identical to those previously used to characterize the products of Tn10 transposition, which were used as a yardstick (6). The gels were stained with SYBR green and recorded on a Fluorimager. Integration events that trap an even number of nodes between the transposon ends and the target site produce catenated deletion circles (Cat), while those that trap an odd number of nodes yield knotted inversion circles (Knot). IC, unknotted inversion circles; ETF, excised transposon fragment; SEST, single-ended-strand-transfer events, yielding lariat structures; 1D, first dimension. The identities of the products were determined by comparison with the gel shown in panel C. Further details are given in the text and reference 6. (C) The transposition reaction and the first dimension of the gel were identical to that in panel B. Annotations are also similar. After electrophoresis in the first dimension, the restriction endonuclease PaeI, which cuts at a single site within the transposon, was diffused into the gel. PaeI digestion converts knotted products into a linear species identical in size to the ETF. Upon digestion, catenated products yield one linear species and one open circular species that together equal the size of the ETF. The relative sizes of the linear and circular species depend on the site of intramolecular integration. Since the gels in panels B and C share a first dimension, the positions of the various products are the same and are indicated by the dashed vertical lines. Further details are given in the text and reference 6.