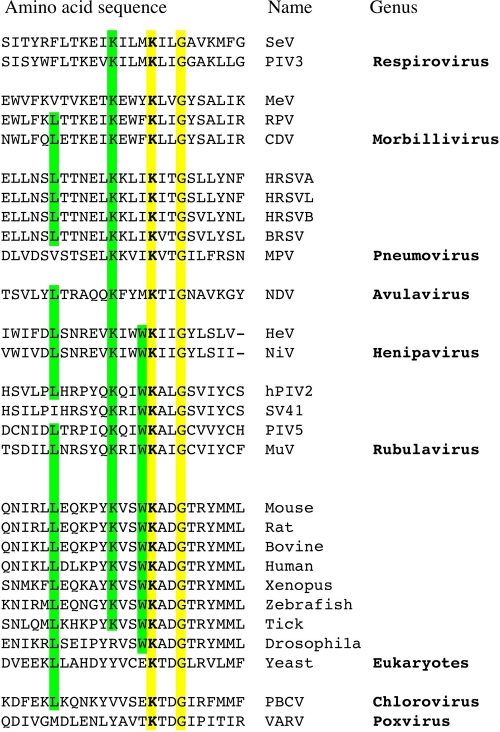
FIG. 6.
Alignment of negative-stranded RNA virus polymerase domains displaying homology with the catalytic site of eukaryotic mRNA guanylyltransferases. The catalytic lysine is marked in bold. SeV, Sendai virus (UniProtKB accession number Q9DUD8); PIV3, parainfluenza virus type 3 (P12577); MeV, measles virus (P12576); RPV, rinderpest virus (P41357); CDV, canine distemper virus (P24658); HRSVA, human respiratory syncytial virus A (strain A2) (P28887); HRSVL, human respiratory syncytial virus A (long strain) (Q9IWW8); HRSVB, human respiratory syncytial virus B (strain B1) (O36635); BRSV, bovine respiratory syncytial virus (O91940); MPV, murine pneumonia virus (Q50EW2); NDV, Newcastle disease virus (Q9DLD3); HeV, Hendra virus (O89344); NiV, Nipah virus (Q997F0); hPIV2, human parainfluenza virus type 2 (P26676); SV41, simian virus 41 (P35341); PIV5, parainfluenza virus type 5 (Q03396); MuV, mumps virus (P30929); mouse (O55236); rat (B5DFA8); bovine (Q2KHX7); human (O60942); Xenopus laevis (Q6NWX1); zebrafish (Q6NY98); tick (B7PBC2); Drosophila melanogaster (Q9VY44); yeast, Saccharomyces cerevisiae (Q01159); PBCV, Paramecium bursaria Chlorella virus (Q84424); VARV, variola major virus (P33057). ScanProsite software (http://www.expasy.org/tools/scanprosite/) was used to scan the experimentally defined C-terminal motif against the UniProt/Swiss-Prot database. The alignment of paramyxovirus polymerases was made by the Muscle method; cellular motifs were added manually.