Abstract
The origin-specific replication of the herpes simplex virus 1 genome requires seven proteins: the helicase-primase (UL5-UL8-UL52), the DNA polymerase (UL30-UL42), the single-strand DNA binding protein (ICP8), and the origin-binding protein (UL9). We reconstituted these proteins, excluding UL9, on synthetic minicircular DNA templates and monitored leading and lagging strand DNA synthesis using the strand-specific incorporation of dTMP and dAMP. Critical features of the assays that led to efficient leading and lagging stand synthesis included high helicase-primase concentrations and a lagging strand template whose sequence resembled that of the viral DNA. Depending on the nature of the minicircle template, the replication complex synthesized leading and lagging strand products at molar ratios varying between 1:1 and 3:1. Lagging strand products (∼0.2 to 0.6 kb) were significantly shorter than leading strand products (∼2 to 10 kb), and conditions that stimulated primer synthesis led to shorter lagging strand products. ICP8 was not essential; however, its presence stimulated DNA synthesis and increased the length of both leading and lagging strand products. Curiously, human DNA polymerase α (p70-p180 or p49-p58-p70-p180), which improves the utilization of RNA primers synthesized by herpesvirus primase on linear DNA templates, had no effect on the replication of the minicircles. The lack of stimulation by polymerase α suggests the existence of a macromolecular assembly that enhances the utilization of RNA primers and may functionally couple leading and lagging strand synthesis. Evidence for functional coupling is further provided by our observations that (i) leading and lagging strand synthesis produce equal amounts of DNA, (ii) leading strand synthesis proceeds faster under conditions that disable primer synthesis on the lagging strand, and (iii) conditions that accelerate helicase-catalyzed DNA unwinding stimulate decoupled leading strand synthesis but not coordinated leading and lagging strand synthesis.
Herpes simplex virus 1 (HSV-1) is the most extensively studied member of the Herpesviridae, a large family of DNA viruses that cause various diseases (41). The virus possesses a double-stranded, linear DNA genome of approximately 153 kbp (14) that circularizes, depending on the stage of infection (15, 22, 35, 45). Although the exact mechanism has not been established yet, HSV-1 appears to replicate the circularized genome using a combination of homologous recombination and rolling circle amplification (40, 43).
HSV-1 encodes seven proteins essential for DNA replication: the heterotrimeric helicase-primase (UL5-UL8-UL52), the heterodimeric DNA polymerase (Pol)-processivity factor complex (UL30-UL42), the single-stranded DNA (ssDNA) binding protein (ICP8), and the origin-binding protein (UL9) (reviewed in references 4, 6, and 31). The helicase-primase complex possesses DNA-dependent ATPase, helicase, and primase activity. Its biological function is to unwind DNA at the replication fork and to synthesize short RNA primers (2 to 14 nucleotides [nt]) for lagging strand synthesis, preferentially at 3′-G-Pyr-Pyr (where Pyr is pyrimidine) initiation sites (38). Under steady-state conditions, primer synthesis is slow and limited by the synthesis of the dinucleotide. The steady-state kcat varies between 0.04 and 0.3 s−1, depending on the base sequence of the initiation site (37). The vast majority of dinucleotides are not extended into primers long enough to be utilized by the herpesvirus polymerase (5, 37). DNA unwinding rates up to 60 to 65 bp s−1 have been measured in vitro, and herpesvirus polymerase did not stimulate the rate of DNA unwinding (12). This rate could accommodate the estimated rate of replication fork movement in vivo (∼50 bp s−1) (1). Although it is possible to classify the UL52 and UL5 subunits based on conserved amino acid motifs as primase (10, 25) and helicase (16, 18), respectively, a complex interdependence exists between the two subunits and both primase and helicase activities require UL5 and UL52 (2, 8). Additionally, whereas DNA-dependent ATPase activity increases linearly with protein concentration, primase activity shows a cooperative dependence on protein concentration with an apparent KD (equilibrium dissociation constant) of around 50 nM. The UL8 subunit does not display any catalytic activity but has been shown to perform various regulatory roles. It stimulates the activities of the UL52-UL5 complex in general and physically interacts with a large number of cellular and herpesvirus proteins (47). Importantly, binding of UL8 to ICP8 forms the basis for helicase stimulation on ICP8-coated DNA templates (11, 19, 42, 46, 48).
The herpesvirus polymerase consists of the UL30 subunit, which harbors the polymerase and 3′ to 5′ exonuclease (exo) activity, and the processivity factor UL42. Unlike conventional DNA sliding clamps, UL42 does not encircle the DNA; rather, it directly binds both UL30 and DNA via basic amino acid residues (7, 20, 26, 49). Using synthetic primer-templates as the substrate, UL30-UL42 incorporates deoxynucleoside triphosphates (dNTPs) at a pre-steady-state rate of 150 s−1 (7, 20, 49), and as for most polymerases, the rate of primer extension is limited by DNA dissociation under steady-state conditions (7). Nucleotide selection by UL30 is fairly inaccurate, averaging 1 misincorporation every 300 copied bases; however, the extension of mismatched primer termini is greatly disfavored (44).
It is not well understood how the HSV replication enzymes interact in the replisome. In most organisms, the polymerases that copy the leading and lagging strand are functionally coupled to synchronize the pace of DNA replication on both strands. The molecular interactions that underlie functional coupling can differ significantly (reviewed in reference 21). The polymerase holoenzymes of both Escherichia coli and bacteriophage T4 are dimeric. In E. coli, the clamp loader complex τ connects two Pol III molecules, whereas disulfide bonds cross-link the leading and lagging strand polymerases of T4. In some regards, the proteins of the T7 replisome resemble the HSV replication machinery: both have monomeric polymerases, processivity factors that do not encircle DNA, and helicase and primase activities that are structurally intertwined. In T7, the helicase-primase (gp4) tethers directly to the processivity factors (thioredoxin) of both replicative polymerases (gp5), and leading strand synthesis pauses every time T7 primase synthesizes an RNA primer (29). However, it is unknown if the HSV replisome functions similarly. It was previously reported that the seven purified herpesvirus replication proteins could perform both leading and lagging strand DNA synthesis on a minicircle template (13), and the synthesis of the two strands appeared to be mechanistically coupled. However, the amount of leading strand product greatly exceeded the amount of lagging strand product, and the cause of this imbalance was unclear.
Recently, we reported that UL30-UL42 extends DNA primer-templates 650- to 26,000-fold more efficiently than RNA primer-templates (5) and that UL30-UL42 does not efficiently utilize the RNA primers synthesized by herpesvirus primase in coupled primase-polymerase assays (<2% utilization). In contrast, human DNA polymerase α does not discriminate between DNA and RNA primers, and supplementing herpesvirus primase-polymerase reactions with Pol α improved the utilization of RNA primers 10- to 20-fold. This observation, in conjunction with the fact that Pol α resides within herpesvirus replication compartments (50), prompted the suggestion that Pol α may play an important role in HSV-1 DNA replication or even be a part of the HSV-1 replisome.
To gain a better understanding of the interaction of the herpesvirus helicase-primase complex with the herpesvirus polymerase and with human Pol α during continuous DNA synthesis, we used synthetic DNA minicircles as templates for coordinated leading and lagging strand synthesis. The DNA sequence and dNTP concentrations were designed to mimic those found in vivo, and protein concentrations were adjusted to optimize activity. Using exonuclease-deficient HSV polymerase (D368A), we observed a 1-to-1 ratio of leading and lagging strand products and used this system to study the effect of minicircle size, ICP8, Mg2+ ions, the 3′ to 5′ exonuclease of UL30, and human Pol α on HSV-1 DNA replication.
MATERIALS AND METHODS
Proteins.
Recombinant baculoviruses were used to express the His9-UL5-UL8-UL52, and the His6-UL42-UL30 complexes. Sf9 insect cells were coinfected with the desired baculoviruses, and the cells were harvested 48 h after infection. The multiplicity of infection was 2:4:4 for the His9-tagged UL5-UL8-UL52 complex and 3:3 for the His6-tagged UL42-UL30 and His6-tagged UL42-UL30 (D368A) complexes. The protein complexes were purified using nickel nitrilotriacetic acid chromatography, as described previously (38). Human DNA polymerase α (His6-p70-p180) and polymerase-primase (p49-p58-His6-p70-p180) were expressed and purified analogously (38). Purified ICP8 was generously provided by Deborah S. Parris (Ohio State University).
Oligonucleotides.
All oligonucleotides were purchased from Integrated DNA Technologies, Inc. Figure 1 depicts the base sequences of the tailed primer (TP) and the 70-base minicircle (MC70). The sequence of the 15-base primer (P) described in Fig. 2 was 5′-TCG GTG GTG CTT GGC. The minicircles were synthesized as follows. For ligation, 1 nmol of 5′phosphorylated 70-base minicircle template was incubated with 1.5 nmol of bridge oligonucleotide 1 (5′-CTG TGT TCC GTC GGT GGT GC) in 12 ml of ligation buffer (50 mM Tris-HCl pH 8, 10 mM MgCl2, 10 mM dithiothreitol [DTT], 1 mM ATP, 0.025 mg/ml bovine serum albumin [BSA]). A total of 120 Weiss units of T4 ligase (USB) was added, and the reaction was allowed to proceed overnight at 16°C. Prior to ethanol (EtOH) precipitation of the minicircle, the reaction mixture was supplemented with EDTA (to 10 mM), NaCl (to 0.3 M), 0.05 mg of glycogen, and 0.01 mg of yeast tRNA (Invitrogen) and extracted twice with phenol-chloroform and twice with chloroform. The resuspended minicircle was gel purified using a 10% polyacrylamide-6 M urea denaturing gel. The template for the 140-base minicircle (MC140) was 5′-Pi-CGG AAC ACA GAA GCG GAA GCA GGG ACA GAA GCG GAA GCA GGG ACA GAA GCC CGG AGC CAA GCA CCA CCG A CGG ACA AAC GGA GAG AAG ACG GAG ACG GCA GCA GAA GAA GAC ACA CAA GGC CAA CGC CGA ACG GAG CAG G, and oligonucleotide bridge 2 (5′ TGC TTC CGC TTC TGT GTT CCG CC TGC TCC GTT CGG CGT TG) was used for ligation.
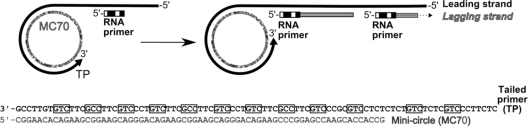
FIG. 1.
Schematic of the rolling-circle mechanism and base sequence of the minicircle substrate. The leading strand is generated by 3′ extension of the tailed primer (TP) using the minicircle (MC70) as a template. The generated leading strand acts as a template for lagging strand synthesis, which is initiated by synthesis of RNA primers. Putative primer initiation sites (3′-G-Pyr-Pyr) are boxed.
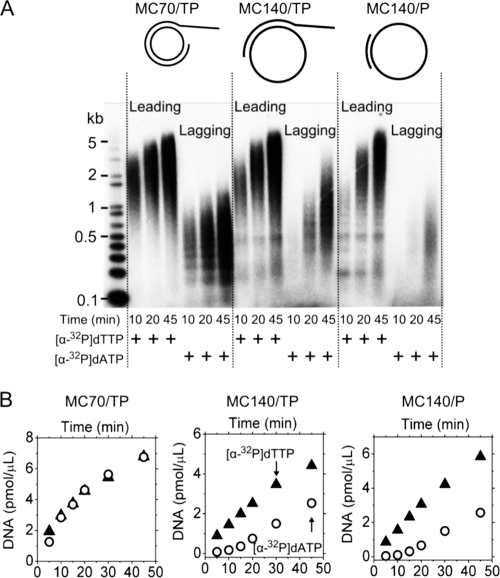
FIG. 2.
DNA synthesis by HSV-1 replication enzymes using different DNA templates. The minicircles are 70 bases (MC70) and 140 bases (MC140) long, and they are hybridized to the tailed primer (TP) or to a simple 15-base oligonucleotide without a 5′ overhang (P). All assays contained 100 nM UL30-UL42, 200 nM UL52-UL5-UL8, 10 nM DNA template, 10 μM each dNTP, 2.8 mM ATP, and 0.8 mM GTP, CTP, and UTP. Leading and lagging strand synthesis was monitored by the strand-specific incorporation of [α-32P]dTTP and [α-32P]dATP, respectively. (A) Separation of the reaction products using alkaline agarose gel electrophoresis. (B) Time courses of leading (▴) and lagging (○) strand synthesis as quantified using filter binding assays. The amount of synthesized DNA is provided as molar concentration of polymerized dNMPs.
Replication assay.
Standard reaction mixtures (10 μl) contained 10 nM DNA, 2.8 mM ATP, 0.8 mM UTP, 0.8 mM CTP, 0.8 mM GTP, 10 μM each dNTP, 100 or 200 nM UL5-UL8-UL52, 100 nM UL42-UL30 (exo deficient [exo−]), 50 mM Tris-HCl (pH 8), 10 mM MgCl2, 1 mM dithiothreitol, 0.1 mg/ml bovine serum albumin, and 5% glycerol. To monitor leading strand synthesis, 1 μCi of [α-32P]dTTP was included, and for lagging strand synthesis 1 μCi of [α-32P]dATP was used. Reaction mixtures with wild-type (wt) UL30 were identical, except for the dNTP concentrations (40 μM dATP, 40 μM dTTP, 100 μM dCTP, 100 μM dGTP) and the use of 8 mM magnesium acetate (MgOAc) instead of 10 mM MgCl2. The reactions were quenched (typically after 45 min) with an equal volume of 150 mM EDTA and analyzed on alkaline agarose gels (1.2% agarose), which were fixed (with 7% trichloroacetic acid) and dried prior to phosphorimaging. Filter binding assays using DE81 paper (Whatman), and three washes with 0.34 M KH2PO4 were employed to quantify the amount of synthesized DNA.
The reactions were started by adding DNA polymerase, while helicase-primase and, if present, ICP8 were added prior to the polymerase (less than 1 min earlier). Note that the herpesvirus helicase-primase appears to be very sensitive to mechanical stress such as aggressive mixing or centrifuging. All data we compare and tabulate in this article were obtained on the same day using the same enzyme aliquots. While 100 or 200 nM helicase-primase gave identical results up to 30 min, at longer times the rate of DNA synthesis declined at 100 nM helicase-primase. Thus, for longer time points we typically used 200 nM helicase-primase.
Primase and coupled primase-polymerase assays.
Primase reaction mixtures (10 μl) contained from 10 nM to 1 μM TP, 0.8 mM ATP, 0.8 mM UTP, 0.8 mM CTP, 0.8 mM GTP, 1 μCi of [α-32P]GTP, and 100 nM UL5-UL8-UL52. Coupled primase-polymerase reaction mixtures additionally contained 100 nM UL30-UL42 (exo−), 10 μM each dNTP, and 1 μCi of [α-32P]dATP. Typically, reactions were stopped after 15 to 45 min by addition of an equal volume of 90% formamide. Primer synthesis was analyzed on denaturing 20% polyacrylamide-8 M urea gels, while primase-coupled polymerase products were analyzed on 12% polyacrylamide-8 M urea gels. The fraction of primers utilized by herpesvirus polymerase was determined by comparing the total moles of product synthesized in the coupled reactions to the total moles of RNA primers (longer than 8 nt long) (5).
Analysis of TMP excision.
Standard reaction mixtures containing 1 μCi of [α-32P]dTTP were stopped with EDTA as described above. From each reaction product 0.5 μl was spotted on a thin-layer chromatography (TLC) plate coated with polyethyleneimine (PEI)-cellulose powder (J. T. Baker, Inc.). A 0.34 M solution of KH2PO4 was used as the liquid phase to separate [α-32P]dTTP from [α-32P]dTMP. The results were visualized using phosphorimaging.
RESULTS
Leading and lagging strand synthesis on different minicircles.
Most of the experiments described in this study were performed using the minicircle substrate depicted in Fig. 1 (MC70/TP). A 70-nt-long DNA molecule lacking thymine was circularized by ligation and hybridized to a 90-nt-long, linear DNA molecule lacking adenine. The resulting partially double-stranded circle has a 40-base single-stranded 5′ tail and a 20-base single-stranded region on the minicircle for protein loading. In addition, it contains a unique HpaII restriction site.
The DNA strand that serves as a template for lagging strand synthesis contains 12 primase initiation sites within 90 nt, which were either 3′-GCC or 3′-GTC. The 3′ G is noncoding, and primer synthesis begins at the next pyrimidine. This frequency of primase initiation sites is only slightly higher than that found in herpesvirus DNA. Since herpesvirus DNA is approximately 67% G·C, primase initiation sites will occur on average once every 12 nt. The leading strand lacks any primer initiation sites. In control experiments, we did not detect any primer synthesis on the leading strand template (data not shown). Since each strand is composed of only 3 of the 4 nucleotides, leading and lagging strand synthesis can be monitored by the strand-specific incorporation of [α-32P]dTTP and [α-32P]dATP, respectively. Initially, we used an exonuclease-deficient form of UL30 (D368A) in the reactions.
We initially examined the ability of the MC70/TP minicircle to support leading and lagging strand synthesis (Fig. 2A). The leading strand was on average 2 to 3 kb long, whereas the average length of the lagging strand was 0.4 kb; the length of both strands increased slightly over time. Figure 2B displays the corresponding time courses as obtained from filter binding assays. At 100 nM polymerase complex and 200 nM helicase-primase, the reaction with MC70/TP produced equal amounts of leading and lagging strand product for the duration of 45 min. Although identical results were obtained using only 100 nM helicase-primase, the rate of DNA synthesis remained constant for a longer period of time using the higher helicase-primase concentration. We used these high concentrations of helicase-primase based on studies showing that primase activity is cooperative, with an apparent KD around 50 nM. Indeed, using less than 100 nM helicase-primase significantly diminished the yield of both leading and lagging strand product. In a typical reaction, about 10% of the available dNTPs were polymerized within 45 min.
We next tested the possibility that the small size of MC70/TP might have affected replication. The persistence length of double-stranded DNA is approximately 50 nm (150 bp), which means that a 70-bp circular duplex is energetically less favorable than the corresponding linear molecule and may therefore induce distortions of the DNA helix. To find out if the small ring size affects the HSV-1 enzymes, we increased the minicircle size to 140 nt (MC140/TP). Since we used the same tailed primer as in MC70/TP, this design also increased the single-stranded gap between the fork and 3′ primer terminus. We did not detect an increase in either the rate of leading strand synthesis or product length. Thus, releasing curvature-induced stress from the double-stranded part of the minicircle did not facilitate the polymerization of nucleotides onto the 3′ terminus of the tailed primer. Notably, though, lagging strand synthesis was time delayed for the larger minicircle and did not catch up to match the amount of leading strand product (Fig. 2B). We attribute this observation to the larger gap between the fork and 3′ primer terminus (20 to 90 bases) which apparently diminishes functional coupling between leading and lagging strand synthesis. Lagging strand synthesis became even less efficient in reactions that employed a completely base-paired 15-nt-long primer that lacked a single-stranded 5′ tail. With this type of primer, the strand displacement activity of the exo− polymerase has to generate the 5′ tail before the helicase-primase can load (53). Using MC70 annealed to the completely base-paired 15-nt-long primer as the substrate resulted in <10% of the amount of leading and lagging strand synthesis observed on MC70/TP (data not shown), providing further evidence for the importance of the single-stranded 5′ tail.
Interdependence between leading and lagging strand synthesis.
In all of the afore described studies with MC70/TP, leading and lagging strand synthesis occurred simultaneously. To determine the characteristics of decoupled leading strand synthesis, we prevented lagging strand synthesis by omitting dATP and/or nucleoside triphosphates (NTPs) in the assays (Fig. 3 A). First, we will discuss the properties of leading strand synthesis. In agreement with previous reports, continuous leading strand synthesis is possible in the absence of NTPs, as long as at least one NTP, preferentially ATP (data not shown), is present to support the ATPase/helicase activity of the helicase-primase complex (33, 53). Adding back the complete set of NTPs, conditions that will allow efficient primer synthesis on the lagging strand template, decreased the amount (∼25% decrease) and length of leading strand product. The stimulation of leading strand synthesis in the absence of GTP, UTP, and CTP was more pronounced at lower Mg2+ ion concentrations (see below), conditions that stimulate the helicase activity. Omitting dATP in the presence of all NTPs allows for primer synthesis on the lagging strand but limits the extension of the primer by dNTPs to the first template T. Although these conditions completely suppressed the formation of lagging strand product, the leading strand product did not lengthen in size. Thus, the rate of leading strand synthesis appears to be limited by both the rate of DNA unwinding and events associated with lagging strand synthesis, the latter being mediated by the complete set of NTPs. In the absence of dTTP, conditions that will prevent leading strand synthesis, lagging strand products were not observed, thereby attesting to the authenticity of the lagging strand signal.
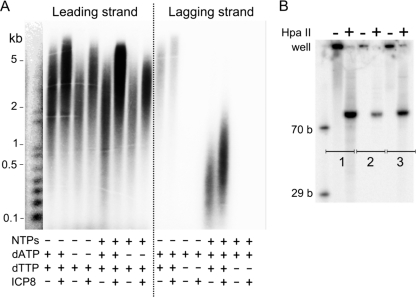
FIG. 3.
Effects of ICP8 and leading or lagging strand suppression on the replication of MC70/TP. (A) Leading strand synthesis was suppressed by omitting dTTPs; lagging strand synthesis was disabled by omitting dATP and/or NTPs. All reaction mixtures contained 10 μM dGTP, 10 μM dCTP, and 2 mM ATP to allow for DNA unwinding. Where indicated, dATP and dTTP were added to a final concentration of 10 μM, and ATP, GTP, CTP, and UTP were added to final concentrations of 2.8, 0.8, 0.8, and 0.8 mM, respectively. ICP8 was added at a concentration of 330 nM. (B) Cleavage of leading and lagging strand products with HpaII. Lanes 1, lagging strand product before (−) and after (+) cleavage; lanes 2, lagging strand product formed in the absence of GTP, UTP, and CTP before and after cleavage; lanes 3, leading strand product before and after cleavage.
We next examined what would happen to lagging strand synthesis under conditions when primer synthesis would be inhibited due to the lack of GTP, CTP, and UTP. In the presence of all dNTPs, [α-32P]dATP, and only ATP (to support helicase activity) (Fig. 3A), a small amount of long radiolabeled product was synthesized, even though primase should not be able to synthesize correctly base-paired RNA primers under these conditions. To decide whether the signal originated from the misinsertion of radiolabeled dATP into the leading strand or if the lagging strand polymerase extended short oligo(rA) primers, we treated the reaction products with HpaII. HpaII will cleave the products only if they are double stranded. Figure 3B shows the leading and lagging strand products (the latter generated in the presence or absence of GTP, UTP, and CTP) before and after treatment with HpaII. The DNA cleavage was nearly complete in all three cases, providing evidence for the prevalence of duplex DNA. While this result was expected for the reaction mixtures that contained all NTPs, it was surprising to find double-stranded lagging strand product in the presence of only ATP. Apparently the primase can synthesize, and the lagging strand polymerase can utilize oligo(rA) primers or oligo(rA) primers that contain misinserted deoxynucleoside monophosphates (dNMPs); however, priming by means of these primers is a rare event and does not lead to balanced leading-to-lagging strand ratios. Rather, efficient lagging strand synthesis strictly required the presence of both ATP and GTP (data not shown), as expected for primer synthesis at the primase initiation sites (i.e., 3′-GTC or 3′-GCC).
Effect of ICP8.
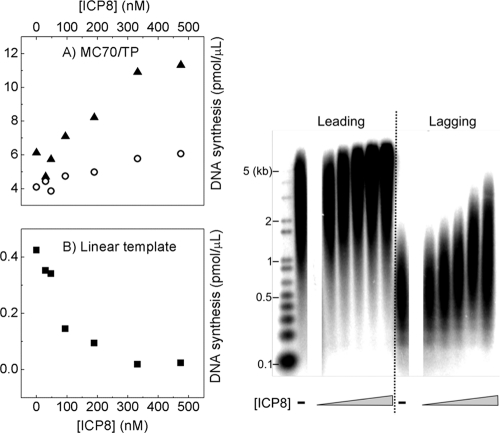
We next examined the effect of ICP8, an essential component of the herpesvirus replication apparatus in vivo that increases the rate of DNA unwinding by purified helicase-primase up to a factor of 30, depending on the Mg2+ ion concentration (12). As we added ICP8 to the MC70/TP reaction mixtures, we observed two effects (Fig. 3A and 4 A). First, the length of the leading and lagging strand products increased under all tested reaction conditions. Second, we observed an ∼2-fold stimulation of leading strand synthesis in the presence of ≥330 nM ICP8, whereas the effect was slightly less pronounced for the lagging strand product. ICP8 had the opposite effect on coupled primase-polymerase reactions on linear templates (TP). The formation of coupled product was completely inhibited at ICP8 concentrations of >200 nM (Fig. 4B). Since one ICP8 molecule binds to roughly 10 nt (17), the 90-nt-long template (at 10 nM) should be completely coated with ICP8 at these higher concentrations. ICP8 has the ability to melt duplex DNA (3), making the displacement of the short and therefore unstable RNA primers a likely explanation for the inhibition of coupled activity on the linear templates. In addition, the herpesvirus polymerase may have difficulties displacing ICP8 molecules that are bound to single-stranded DNA upstream from it.
FIG. 4.
Effects of ICP8 on leading and lagging strand synthesis. Comparison of the effect of ICP8 on rolling-circle amplification (A) and on coupled primase-polymerase activity using the tailed primer alone as a template (B). The gels on the right show the size distribution of the products. The assay conditions were identical, except for the nature of the DNA templates (10 nM MC70/TP and 10 nM TP, respectively). In panel A, triangles indicate leading strand products, and circles indicate lagging strand products. Panel B shows DNA synthesized on the linear template.
Mg2+ ion dependence of ICP8 stimulation.
The fact that ICP8 stimulates the replication of minicircles at concentrations that inhibit the coupled action of primase and polymerase on linear templates suggests that an interaction between ICP8 and the helicase is responsible for the stimulation of minicircle replication. Characteristic for the herpesvirus helicase activity in the presence of ICP8 is its dependence on Mg2+ ion concentration: decreasing the MgCl2 concentration from 4 to 1 mM dramatically increases the rate of the unwinding of ICP8-coated DNA (12). To determine if the ICP8-catalyzed minicircle replication shares this characteristic, we studied the effect of MgCl2 concentration on DNA synthesis.
We tested the hypothesis that ICP8 would differentially affect coupled leading and lagging strand synthesis versus uncoupled leading strand synthesis. Leading and lagging strand synthesis in the presence of ICP8 was insensitive to varying the MgCl2 concentration between 7 and 10 mM (corresponding roughly to 1.8 and 4.8 mM free Mg2+ ions at an NTP [total] concentration of 5.2 mM) (Fig. 5 A). However, when we omitted all NTPs but ATP from the reaction mixture, thus inhibiting primer synthesis, we observed an increase of leading strand product upon lowering the MgCl2 concentrations (Fig. 5A). Thus, in the absence of primer synthesis, the rate of leading strand synthesis follows the same trend previously reported for helicase-catalyzed DNA unwinding, suggesting that DNA unwinding is rate limiting during decoupled leading strand synthesis. The absence of an effect of Mg2+ during coupled leading and lagging strand synthesis, however, suggests that some process other than helicase unwinding is now rate limiting. The effect of Mg2+ on the wild-type (exo+) herpesvirus polymerase is discussed below (Fig. 5A and B).
FIG. 5.
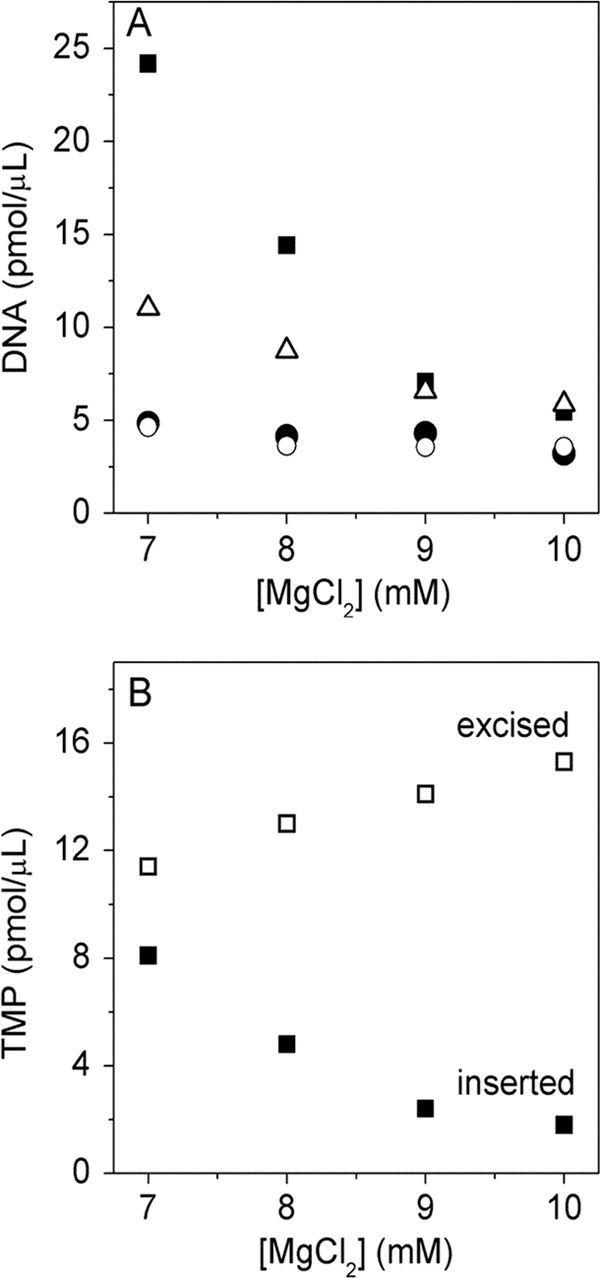
Effect of MgCl2 concentration on DNA synthesis. (A) Leading and lagging strand synthesis at different MgCl2 concentrations in the presence of 330 nM ICP8 using MC70/TP as a template. Depicted are standard reactions employing HSV exo− polymerase (•, leading strand; ○, lagging strand) or wild-type HSV polymerase (▪, leading strand) and exo− polymerase in the absence of GTP, CTP, and UTP (▵, leading strand). Note that the latter two reactions produced little or no lagging strand product; thus, they represent decoupled DNA synthesis. (B) Analysis of the TMPs excised (□) or inserted (▪) by the wild-type polymerase as a function of MgCl2 concentration.
Rates of coupled and decoupled DNA synthesis.
To provide a more quantitative understanding of replication and compare the minicircle system to what happens in vivo, we measured the rates of leading and lagging strand chain elongation. Adding ICP8 or omitting CTP, GTP, and UTP stimulated leading strand synthesis. To find out if the increased yield of DNA is due to faster polymerization rates, changes in polymerase processivity or perhaps a more efficient turnover of minicircles, we analyzed the length of the DNA products at early times. In particular, we compared the length of leading strand products synthesized during coordinated leading and lagging strand synthesis (coupled reaction) as well as the effects of suppressing primer synthesis on the lagging strand (decoupled reaction). DNA products were detectable by gel electrophoresis after a reaction time of 2.5 min (Fig. 6; because of their vastly different intensities, we omitted the 2.5 min data point from the gel). Similar to the results at longer times (see above), the leading strand synthesized during coupled DNA synthesis was shorter than the DNA product obtained in the decoupled reaction, and ICP8 increased the length in both cases. The lengths of the leading strand products at 5 and 10 min provide a rough estimate of the polymerization rates. Without ICP8, the rates of the coupled and decoupled reactions are 4 and 6 s−1, respectively, and with ICP8, 8 and 13 s−1, respectively. Thus, the replication fork proceeds around 50% faster if no RNA primers are synthesized, and ICP8 accelerates the fork movement by a factor of 2.
FIG. 6.
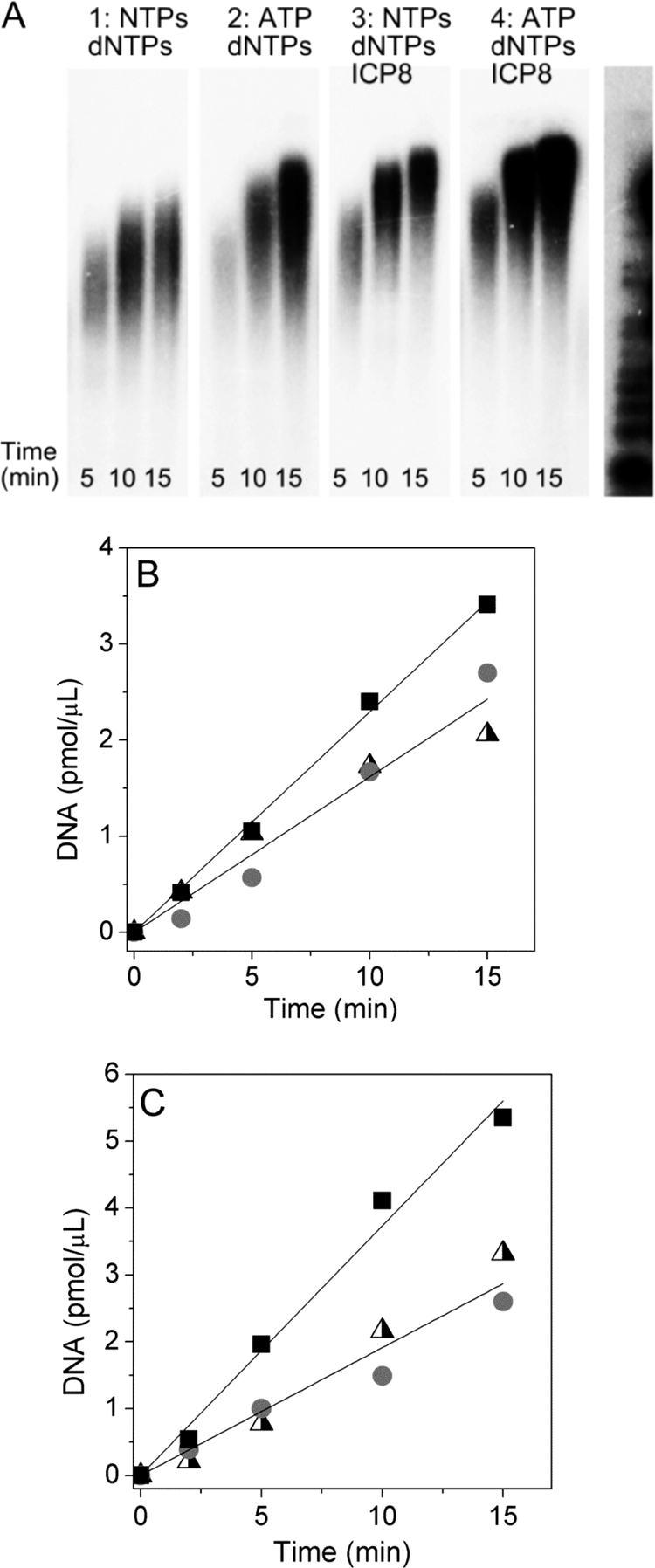
Rates of DNA synthesis. (A) Length of leading strand products as a function of time. Coupled reaction mixtures contained 100 nM UL30-UL42, 200 nM UL52-UL5-UL8, 10 nM MC70/TP, 7 mM MgCl2, 10 μM each dNTP, 2.8 mM ATP, and 0.8 mM GTP, CTP, and UTP. Decoupled reactions lacked GTP, CTP, and UTP. Where indicated, 330 nM ICP8 was included.(B) Amount of DNA synthesized in reaction mixtures without ICP8. (C) Amount of DNA synthesized in reaction mixtures containing ICP8. Decoupled reaction, squares; coupled reactions, triangles (leading strand) and circles (lagging strand) (B and C).
Lack of stimulation by human DNA polymerase α.
Previous work showed that herpesvirus polymerase uses RNA primers extremely inefficiently, unlike the cellular DNA Pol α that elongates RNA primers very efficiently (5). Additionally, herpesvirus replication compartments contain Pol α (50). These data raised the possibility that Pol α might improve the utilization of RNA primers synthesized by herpesvirus primase and thereby stimulate replication. Adding 2 to 80 nM Pol α had no effect on the outcome of the minicircle reactions (Fig. 7 B). To exclude the possibility that the lack of Pol α stimulation was due to the base sequence of the minicircle substrate, we performed coupled primase-polymerase reactions using 1 μM linear TP as the template. In the presence of 100 nM herpesvirus helicase-primase and 100 nM herpesvirus polymerase, 20 nM Pol α increased the yield of long DNA product by a factor of 3 (Fig. 7A), similar to previous studies showing that human Pol α enhances the elongation of herpesvirus primase-synthesized primers (5). Thus, the inability of Pol α to stimulate DNA replication on the minicircle was not due to an inability of Pol α to elongate the herpesvirus primase-synthesized primers on this template.
FIG. 7.
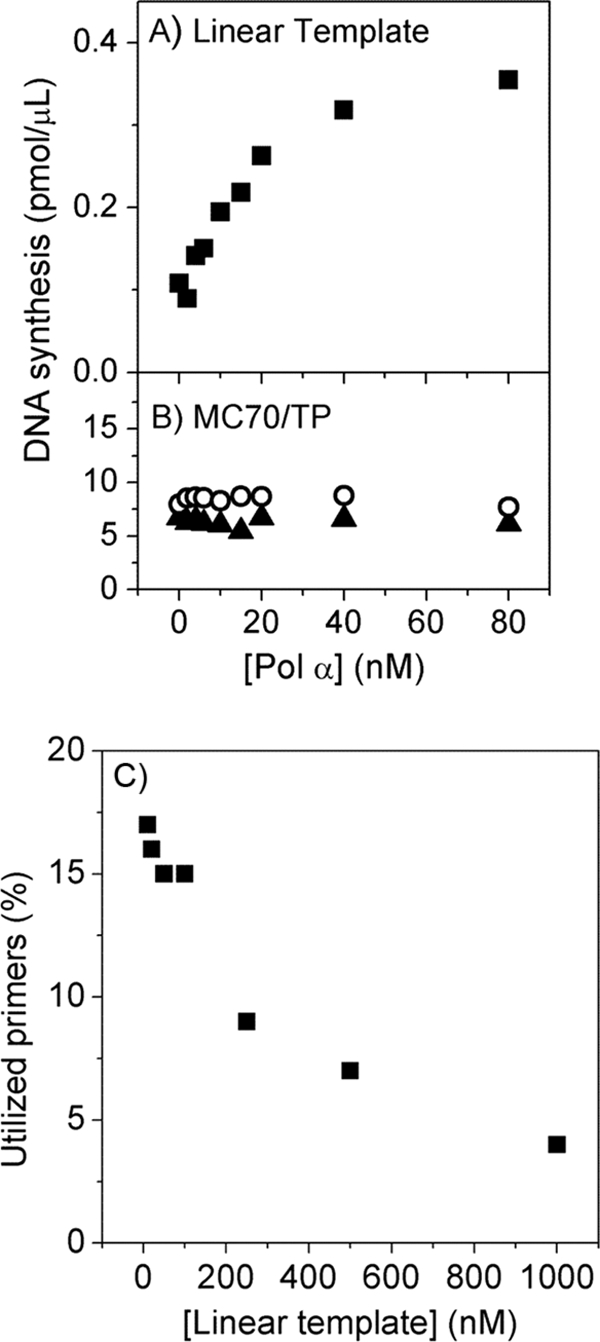
Lack of stimulation of rolling-circle amplification by human DNA polymerase α. (A) Coupled primase-polymerase assays were performed on the linear template as described in Materials and Methods. (B) Replication assays on MC70/TP were performed and quantified as described in Materials and Methods (○, lagging strand; ▴, leading strand). (C) Fraction of the primase-synthesized primers elongated by herpesvirus polymerase on the linear template (TP).
We next sought to explain the dichotomy between the inefficient utilization of RNA primers by herpesvirus polymerase and the apparently efficient utilization of primase-synthesized primers in the minicircle system. In addition to the nature of the DNA substrates, the assays described in Fig. 7A and B differed in the template concentrations used. The minicircle reaction mixtures contained only 10 nM MC70/TP, whereas the concentration of linear template was 1 μM. To determine if the template concentration affects primer utilization by herpesvirus polymerase, we measured primer utilization as a function of template concentration. Remarkably, the fraction of utilized primers increased from 3% at 1 μM TP to 17% at 10 nM TP. Thus, high template concentrations interfere with the ability of UL30-UL42 to use primase-synthesized primers.
Minicircle replication using wild-type (exo+) polymerase.
We tested whether the minicircle system could tolerate replacement of the exo− UL30 with wild-type UL30. However, the replication assays yielded only small amounts of leading strand product and no lagging strand product even though they contained 40 μM concentrations of the dNTPs to help accommodate the robust exonuclease activity of wild-type UL30 (see below). Conducting the reactions at a lower MgCl2 concentration improved leading strand synthesis (3 pmol/μl at 7 mM MgCl2) but did not recover lagging strand synthesis. Similar to the exo− Pol, leading strand synthesis in the presence of ICP8 but in the absence of GTP, UTP, and CTP (see above) depended strongly on MgCl2 concentration (Fig. 5A).
We restored lagging strand synthesis by replacing the MgCl2 with 8 mM Mg(OAc)2. Under these conditions the exo+ enzyme polymerized leading and lagging strand product at ratios varying between 2:1 and 3:1 (Table 1). Similar ratios were obtained for the exo− Pol in the presence of Mg(OAc)2 (Table 1). The leading and lagging strand products synthesized by wt and exo− polymerase in the presence of Mg(OAc)2 were longer than those synthesized in the presence of MgCl2; the leading strand was ∼5 kb, and the lagging strand was ∼1.5 kb long (Fig. 8 A). The addition of ICP8 increased the length of both strands further. The increased length of lagging strand products was not a consequence of the elevated dNTPs since replacing the Mg(OAC)2 with MgCl2 in reaction mixtures containing the exo− UL30 restored the shorter lagging strand products (Fig. 8B). Thus, the Mg(OAC)2 both allows lagging strand synthesis to occur with the wild-type UL30 and also increases the length of the Okazaki fragments.
TABLE 1.
Amount of DNA produced in 45 min in assays containing 10 nM of the designated DNA template and MgOAc or MgCl2a
| DNA template | Treatment | MC70/TP replication |
MC140/TP replication |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Leading strand (pmol/μl) |
Lagging strand (pmol/μl) |
Leading strand (pmol/μl) |
Lagging strand (pmol/μl) |
||||||
| No ICP8 | With ICP8 | No ICP8 | With ICP8 | No ICP8 | With ICP8 | No ICP8 | With ICP8 | ||
| HSV exo+ | MgOAc | 5.2 | 8.4 | 1.7 | 2.5 | 4.7 | 12 | 3.1 | 2.7 |
| HSV exo− | MgOAc | 8.8 | 16 | 4.8 | 11 | 9.1 | 17 | 4.1 | 7.2 |
| HSV exo− | MgCl2 | 6.2 | - | 6.7 | - | ||||
Mixtures also contained the following: 40 μM dATP and dTTP; 100 μM dGTP; 100 μM dCTP; 3 mM ATP; 1 mM GTP, CTP, and UTP; 8 mM MgOAc or 10 mM MgCl2; 100 nM UL30-UL42; 200 nM UL52-UL5-UL8; and 330 nM ICP8.
FIG. 8.
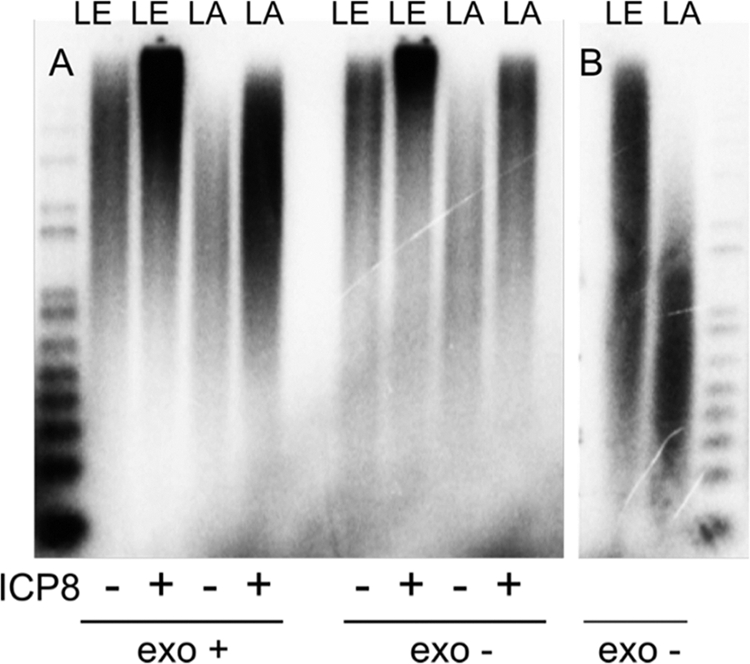
Leading (LE) and lagging (LA) strand synthesis with wild-type and exo− polymerase. (A) All assays contained 10 nM MC70/TP, 8 mM Mg(OAc)2, 100 nM UL30-UL42, 200 nM UL5-UL8-UL52, 40 μM dATP and dTTP, 100 μM dGTP, 100 μM dCTP, 2.8 mM ATP, and 0.8 mM GTP, CTP, and UTP. The ICP8 concentration was 330 nM. (B) The reaction mixtures contained 10 mM MgCl2 instead of the Mg(OAc)2 to determine if the longer lagging strand products are solely due to the higher dNTP concentrations or are also due to the altered salt conditions.
Wild-type UL30 contains a highly active 3′ to 5′exonuclease; hence, it seemed likely that this activity would excise substantial amounts of newly incorporated nucleotides as dNMPs. Remarkably, more TTP is converted into TMP than is stably incorporated into DNA (Fig. 5B). To better understand why so much TMP is produced, we examined the effects of increasing the rate of DNA synthesis. Under conditions where only leading strand synthesis can occur, decreasing the Mg2+ concentration increases the rate of DNA synthesis, likely by increasing the rate of DNA unwinding (12) (Fig. 5A). Figure 5B shows that this also results in decreased conversion of TTP to TMP.
Inhibition of replication.
To both determine if the minicircle system could ultimately be used for analyzing the effects of inhibitors and demonstrate that the system responds appropriately to a known inhibitor of herpesvirus replication, we supplemented the minicircle reaction mixture with the herpesvirus polymerase inhibitor acyclovir triphosphate. Acyclovir is a sugar analogue of guanosine, where the sugar ring is replaced by an open chain structure that lacks the 2′ and 3′ carbons. Herpesvirus polymerase inserts acyclovir triphosphate readily and disables DNA replication due to chain termination. Consistent with the mechanism of action, acyclovir triphosphate inhibited leading and lagging strand synthesis equally with a 50% inhibitory concentration (IC50) of 7 μM (Fig. 9).
FIG. 9.
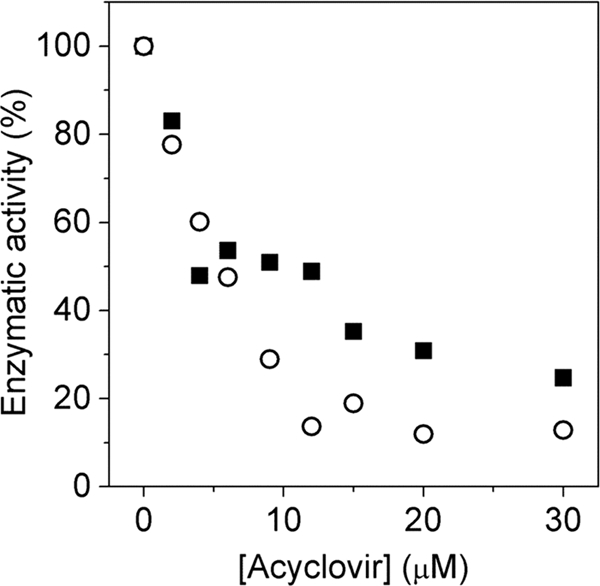
Inhibition of leading (▪) and lagging (○) strand synthesis using acyclovir triphosphate.
DISCUSSION
In this report, we used synthetic DNA minicircles to investigate the mechanism of leading and lagging strand synthesis by HSV replication enzymes. The reconstituted replisome, consisting of the helicase-primase and the processivity factor-polymerase (exo−) complex, synthesized leading and lagging strand products with similar rates. The resulting DNA was double stranded, pointing toward a coordinated mechanism of DNA synthesis. Although the coordinated leading and lagging strand replication of minicircular DNA has been reported for replication complexes of E. coli (32, 51) and of bacteriophages T4 (23, 39) and T7 (28), it had not been observed previously for any eukaryotic systems.
The similar rates of leading and lagging strand synthesis reported herein contrast with the previous report that showed a very large excess of leading strand products over lagging strand products (13). This likely resulted from three significant changes. (i) We used concentrations of NTPs and dNTPs similar to those found in vivo. Previous work used higher concentrations of dNTPs and lower concentrations of NTPs, conditions that can result in significant primase inhibition (24). (ii) We used higher concentrations of UL5-UL8-UL52 (100 nM) to enhance primase activity. (iii)Our minicircle had a primase initiation site (3′-G-Pyr-Pyr) approximately every 8 nucleotides and was G-C rich (65%), similar to the situation in HSV-1 DNA (a primase initiation site approximately every 12 nucleotides and 68% G-C). In contrast, previous minicircles contained one initiation site among 70 nucleotides.
Without ICP8, the rate of DNA replication was ∼12 times lower than the speed of replication fork movement in vivo (50 bp s−1 [1]) and as fast as the replication of primed M13mp6 DNA by HSV-infected Vero cell extracts (36). Although the single-strand DNA binding protein ICP8 was not strictly required, its presence increased the rate of DNA polymerization by a factor of 2. The lower rates of fork movement using a minimal set of proteins likely indicate that optimal replication rates require additional proteins, either cellular or herpesvirus encoded, and that the buffer composition used for our studies varies substantially from what is found in vivo. In vivo, ICP8 is essential for HSV-1 replication, and a recent proteomics study identified about 50 cellular proteins that interact with ICP8 in HSV-1 replication compartments (47). While it is unclear which of these cellular proteins is important, the large number of molecular interactions emphasizes the importance of ICP8 as a mediator protein.
On average, the leading and lagging strand products were around 2.5 kb and 0.4 kb long, respectively, and as long as ≥10 kb and 1 kb in the presence of ICP8. The length of the lagging strand product compares well with Okazaki fragments synthesized on minicircle templates by T7 (1 kb) (28) and T4 (1 to 2 kb) (23, 39) and in vivo by E. coli (1 to 2 kb) and eukaryotic cells (0.1 to 0.2 kb) (34). As observed for E. coli, T7, and T4, the synthesis of the Okazaki fragments depended on the NTP pool and, hence, on primase activity (30, 51, 52). For herpesvirus primase, the presence of ATP and GTP was strictly required for the synthesis of short Okazaki fragments, but the length was fairly insensitive to the NTP concentration, at least within the examined range (>250 μM). Even under conditions that stimulate primase activity (i.e., high NTP concentrations), the length of the Okazaki fragments did not fall below 0.3 kb, despite the abundance of primer initiation sites on the lagging strand template (on average, 1 priming site every 8 nt). Notably, adding ICP8 increased the length of the Okazaki fragments in parallel with the rate of leading strand synthesis. This suggests that the length of the Okazaki fragment is coupled to the rate of leading strand synthesis and controlled by mechanisms other than the mere availability of priming sites and NTPs. It is also interesting that balanced ratios of leading and lagging strand always coincided with relatively short Okazaki fragments, as if frequent priming was a result of functional coupling between leading and lagging strand synthesis.
Unlike Kadyrov and Drake, who reported a 7-fold increase in polymerization rate for reconstituted T4 proteins upon increasing the minicircle size (from 70 to 120 bp) (23), the minicircle size did not affect the polymerization rate or product length in the herpesvirus system. However, the nature of the protein loading site appears to be very important for coordinated DNA synthesis in the herpesvirus system. Using the DNA minicircle that exhibited a 20-base-long single-stranded region between the 3′ terminus of the leading strand primer and the fork junction, we observed optimally synchronized lagging strand synthesis. However, when the single-stranded region was 90 bases long, the onset of lagging strand synthesis was delayed by 10 to 15 min. Likewise, simply annealing a primer to the minicircle resulted in a distinct lag in lagging strand synthesis and less total lagging strand product. These results suggest that single-stranded regions neighboring the replication fork play critical roles for efficiently assembling a replisome. Experiments to better define these requirements are in progress.
A process associated with lagging strand synthesis likely restricts the overall rate of DNA replication in the minicircle system. A unique characteristic of the herpesvirus helicase activity in the presence of ICP8 is its dependence on Mg2+ ion concentration. Disconnected from the replisome, the helicase unwinds ICP8-coated DNA with a maximal rate at 1 mM MgCl2 (12) while higher concentrations inhibit activity. To find out if ICP8 stimulates minicircle replication by increasing the rate of DNA unwinding, we varied the free Mg2+ concentrations between 1 and 4 mM. While the coordinated leading and lagging strand DNA synthesis in the presence of ICP8 was insensitive to Mg2+ concentration, leading strand synthesis in the absence of lagging strand synthesis was strongly Mg2+ ion dependent. This finding suggests that the functional coupling between leading and lagging strand synthesis sets an upper limit to replication fork movement that is not exceeded, even under conditions that stimulate the helicase. Interestingly, omitting dATP, the one dNTP essential for lagging strand synthesis but not needed for leading strand synthesis, in the presence of all NTPs did not accelerate leading strand synthesis. Thus, primer synthesis itself and/or the addition of just a few dNTPs onto the primer, and not the full extension of RNA primers, appeared to mediate functional coupling between the leading and lagging strand polymerase.
The view that the helicase-primase and polymerase adopt a special mode of interaction during minicircle replication is also supported by the lack of lagging strand stimulation by human Pol α. Since Pol α elongates RNA primers much more efficiently than the herpesvirus polymerase, it should quickly bind and extend vacant RNA primers (5). This is the mechanism by which Pol α improves the utilization of primers synthesized by the coupled action of herpesvirus primase and polymerase on linear templates. The absence of a Pol α effect suggests that herpesvirus polymerase uses the primers synthesized during minicircle replication more efficiently than on a linear template.
Herpesvirus polymerase elongates primase-synthesized primers more efficiently at low template concentrations than at high template concentrations. At 10 nM linear template, the polymerase elongated 17% of the primase-synthesized primers that were at least 8 nt long. While this is much higher than previously reported (1 to 2% [5]), this fraction is still remarkably low, given that the assay contained excess polymerase (100 nM). A likely explanation for the enhanced primer utilization at low DNA concentrations is that excess single-stranded DNA cannot bind to UL30 and compete with binding of the RNA primer-template.
Obtaining both leading and lagging strand synthesis by the wild-type (exo+) herpesvirus polymerase required increasing the dNTP concentration to 40 μM and replacing the MgCl2 with Mg(OAc)2. Compared to replication using the exo− polymerase, both leading and lagging strand products increased in length, and somewhat more leading strand products were obtained. The slightly imbalanced leading-to-lagging-strand ratio (around 2 to 3:1) was largely a consequence of using Mg(OAc)2 since using Mg(OAc)2 with the exo− Pol also now resulted in imbalanced synthesis. Why changing the anion from Cl− to OAc− had such a large effect remains unclear.
Despite the less balanced ratio of leading and lagging strand, two data indicate functional coupling. As with the exo− Pol, leading strand synthesis was faster (4-fold) when primer synthesis on the lagging strand should not occur (i.e., in the absence of CTP, GTP, and UTP), and Pol α did not increase the yield of lagging strand products (data not shown). Interestingly, the lagging strand products were slightly longer with the exo+ Pol than with the exo− enzyme. This may result from the exonuclease hydrolyzing some of the primase-synthesized primers, thus resulting in fewer initiation sites for synthesizing lagging strand product.
The amount of dNMPs that was incorporated into DNA and then excised by the intrinsic exonuclease activity of UL30 equaled or even exceeded the amount of dNMPs the polymerase inserted into DNA. The catalytic subunits of T7 and the HSV-1 polymerase have very potent exonuclease activities (7.9 s−1 [9] and 5.9 s−1 [7], respectively) compared to other polymerases, such as Klenow fragment (0.002 s−1 [27]). In idling turnover mode, the UL30-UL42 complex converts dNTPs into dNMPs at a rate of 0.9 s−1 (53). The relatively low rate of fork movement (4 to 13 s−1) may contribute to the remarkably large amount of dNMPs produced since it provides the exonuclease a greater opportunity to excise a just-added nucleotide. After a nucleotide is incorporated, either the polymerase activity can add the next required nucleotide, or the exonuclease activity can remove a dNMP. During processive synthesis on linear primer-templates, the isolated herpesvirus polymerase elongates primers with rates ranging between 27 (49) and 44 nt s−1 (7). Conditions that reduce the rate of dNTP polymerization (e.g., slow fork movement) will, therefore, enhance the removal of dNMPs. Consistent with this hypothesis, more dNMP is produced at high Mg2+ concentrations. At low Mg2+ concentrations, the helicase reaches maximal speed, and the polymerase follows without stalling, thus excising fewer dNMPs.
These studies have developed a coupled leading and lagging strand synthesis system for herpesvirus replication. Key features of the system include the relatively high protein concentrations and a lagging strand template that contains frequent initiation sites for herpesvirus primase, similar to herpesvirus DNA. Importantly, this system now provides a basis from which to ask how other herpesvirus and cellular proteins affect replication and for developing systems that mimic either origin-dependent replication or recombination-dependent replication.
Acknowledgments
This work was supported by NIH grants AI59764 and GM073832.
Footnotes
Published ahead of print on 10 November 2010.
REFERENCES
- 1.Ben-Porat, T., M. L. Blankenship, J. M. Demarchi, and A. S. Kaplan. 1977. Replication of herpes virus DNA. III. Rate of DNA elongation. J. Virol. 22:734-741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Biswas, N., and S. K. Weller. 1999. A mutation in the C-terminal putative Zn+2 finger motif of UL52 severely affects the biochemical activities of the HSV-1 helicase-primase subcomplex. J. Biol. Chem. 274:8068-8076. [DOI] [PubMed] [Google Scholar]
- 3.Boehmer, P. E., and I. R. Lehman. 1993. Herpes-simplex virus type-1 ICP8-helix-destabilizing properties. J. Virol. 67:711-715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boehmer, P. E., and I. R. Lehman. 1997. Herpes simplex virus DNA replication. Annu. Rev. Biochem. 66:347-384. [DOI] [PubMed] [Google Scholar]
- 5.Cavanaugh, N. A., and R. D. Kuchta. 2009. Initiation of new DNA strands by the herpes simplex virus-1 primase-helicase complex and either herpes DNA polymerase or human DNA polymerase alpha. J. Biol. Chem. 284:1523-1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chattopadhyay, S., Y. Chen, and S. K. Weller. 2006. The two helicases of herpes simplex virus type 1 (HSV-1). Front. Biosci. 11:2213-2223. [DOI] [PubMed] [Google Scholar]
- 7.Chaudhuri, M., L. Song, and D. S. Parris. 2003. The herpes simplex virus type 1 DNA polymerase processivity factor increases fidelity without altering pre-steady-state rate constants for polymerization or excision. J. Biol. Chem. 278:8996-9004. [DOI] [PubMed] [Google Scholar]
- 8.Chen, Y., S. D. Carrington-Lawrence, P. Bai, and S. K. Weller. 2005. Mutations in the putative zinc-binding motif of UL52 demonstrate a complex interdependence between the UL5 and UL52 subunits of the human herpes simplex virus type 1 helicase/primase complex. J. Virol. 79:9088-9096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Donlin, M. J., S. S. Patel, and K. A. Johnson. 1991. Kinetic partitioning between the exonuclease and polymerase sites in DNA error correction. Biochemistry 30:538-546. [DOI] [PubMed] [Google Scholar]
- 10.Dracheva, S., E. V. Koonin, and J. J. Crute. 1995. Identification of the primase active site of the herpes simplex virus type 1 helicase-primase. J. Biol. Chem. 270:14148-14153. [DOI] [PubMed] [Google Scholar]
- 11.Falkenberg, M., D. A. Bushnell, P. Elias, and I. R. Lehman. 1997. The UL8 subunit of the heterotrimeric herpes simplex virus type 1 helicase-primase is required for the unwinding of single strand DNA-binding protein (ICP8)-coated DNA substrates. J. Biol. Chem. 272:22766-22770. [DOI] [PubMed] [Google Scholar]
- 12.Falkenberg, M., P. Elias, and I. R. Lehman. 1998. The herpes simplex virus type 1 helicase-primase. Analysis of helicase activity. J. Biol. Chem. 273:32154-32157. [DOI] [PubMed] [Google Scholar]
- 13.Falkenberg, M., I. R. Lehman, and P. Elias. 2000. Leading and lagging strand DNA synthesis in vitro by a reconstituted herpes simplex virus type 1 replisome. Proc. Natl. Acad. Sci. U. S. A. 97:3896-3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fields, B. N., D. M. Knipe, P. M. Howley, R. M. Chanock, J. L. Melnick, T. P. Monath, B. Roizman, and S. E. Straus (ed.). 1996. Fields virology, 3rd ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 15.Garber, D. A., S. M. Beverly, and D. M. Coen. 1993. Demonstration of circularization of herpes-simplex virus-DNA following infection using pulsed-field gel-electrophoresis. Virology 197:459-462. [DOI] [PubMed] [Google Scholar]
- 16.Gorbalenya, A. E., E. V. Koonin, A. P. Donchenko, and V. M. Blinov. 1988. A conserved NTP-motif in putative helicases. Nature 333:22. [DOI] [PubMed] [Google Scholar]
- 17.Gourves, A. S., N. Tanguy Le Gac, G. Villani, P. E. Boehmer, and N. P. Johnson. 2000. Equilibrium binding of single-stranded DNA with herpes simplex virus type I-coded single-stranded DNA-binding protein, ICP8. J. Biol. Chem. 275:10864-10869. [DOI] [PubMed] [Google Scholar]
- 18.Graves-Woodward, K. L., and S. K. Weller. 1996. Replacement of gly815 in helicase motif V alters the single-stranded DNA-dependent ATPase activity of the herpes simplex virus type 1 helicase-primase. J. Biol. Chem. 271:13629-13635. [DOI] [PubMed] [Google Scholar]
- 19.Hamatake, R. K., M. Bifano, W. W. Hurlburt, and D. J. Tenney. 1997. A functional interaction of ICP8, the herpes simplex virus single-stranded DNA-binding protein, and the helicase-primase complex that is dependent on the presence of the UL8 subunit. J. Gen. Virol. 78:857-865. [DOI] [PubMed] [Google Scholar]
- 20.Hamatake, R. K., M. Bifano, D. J. Tenney, W. W. Hurlburt, and M. G. Cordingley. 1993. The herpes-simplex virus type-1 DNA polymerase accessory protein, UL42, contains a functional protease-resistant domain. J. Gen. Virol. 74:2181-2189. [DOI] [PubMed] [Google Scholar]
- 21.Hamdan, S. M., and C. C. Richardson. 2009. Motors, switches, and contacts in the replisome. Annu. Rev. Biochem. 78:205-243. [DOI] [PubMed] [Google Scholar]
- 22.Jackson, S. A., and N. A. DeLuca. 2003. Relationship of herpes simplex virus genome configuration to productive and persistent infections. Proc. Natl. Acad. Sci. U. S. A. 100:7871-7876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kadyrov, F. A., and J. W. Drake. 2002. Characterization of DNA synthesis catalyzed by bacteriophage T4 replication complexes reconstituted on synthetic circular substrates. Nucleic Acids Res. 30:4387-4397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Keller, K. E., N. Cavanaugh, and R. D. Kuchta. 2008. Interaction of herpes primase with the sugar of a NTP. Biochemistry 47:8977-8984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klinedinst, D. K., and M. D. Challberg. 1994. Helicase-primase complex of herpes simplex virus type 1: a mutation in the UL52 subunit abolishes primase activity. J. Virol. 68:3693-3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Komazin-Meredith, G., W. L. Santos, D. J. Filman, J. M. Hogle, G. L. Verdine, and D. M. Coen. 2008. The positively charged surface of herpes simplex virus UL42 mediates DNA binding. J. Biol. Chem. 283:6154-6161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuchta, R. D., P. Benkovic, and S. J. Benkovic. 1988. Kinetic mechanism whereby DNA polymerase-1 (Klenow) replicates DNA with high fidelity. Biochemistry 27:6716-6725. [DOI] [PubMed] [Google Scholar]
- 28.Lee, J., P. D. Chastain II, T. Kusakabe, J. D. Griffith, and C. C. Richardson. 1998. Coordinated leading and lagging strand DNA synthesis on a minicircular template. Mol. Cell 1:1001-1010. [DOI] [PubMed] [Google Scholar]
- 29.Lee, J. B., R. K. Hite, S. M. Hamdan, X. S. Xie, C. C. Richardson, and A. M. van Oijen. 2006. DNA primase acts as a molecular brake in DNA replication. Nature 439:621-624. [DOI] [PubMed] [Google Scholar]
- 30.Lee, J., P. D. Chastain II, J. D. Griffith, and C. C. Richardson. 2002. Lagging strand synthesis in coordinated DNA synthesis by bacteriophage T7 replication proteins. J. Mol. Biol. 316:19-34. [DOI] [PubMed] [Google Scholar]
- 31.Lehman, I. R., and P. E. Boehmer. 1999. Replication of herpes simplex virus DNA. J. Biol. Chem. 274:28059-28062. [DOI] [PubMed] [Google Scholar]
- 32.McInerney, P., and M. O'Donnell. 2004. Functional uncoupling of twin polymerases: mechanism of polymerase dissociation from a lagging-strand block. J. Biol. Chem. 279:21543-21551. [DOI] [PubMed] [Google Scholar]
- 33.Nimonkar, A. V., and P. E. Boehmer. 2004. Role of protein-protein interactions during herpes simplex virus type 1 recombination-dependent replication. J. Biol. Chem. 279:21957-21965. [DOI] [PubMed] [Google Scholar]
- 34.Ogawa, T., and T. Okazaki. 1980. Discontinuous DNA replication. Annu. Rev. Biochem. 49:421-457. [DOI] [PubMed] [Google Scholar]
- 35.Poffenberger, K. L., and B. Roizman. 1985. A nonconverting genome of a viable herpes simplex virus 1: presence of head-to-tail linkages in packaged genomes and requirements for circularization after infection. J. Virol. 53:587-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rabkin, S. D., and B. Hanlon. 1990. Herpes simplex virus DNA synthesis at a preformed replication fork in vitro. J. Virol. 64:4957-4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ramirez-Aguilar, K. A., and R. D. Kuchta. 2004. Mechanism of primer synthesis by the herpes simplex virus 1 helicase-primase. Biochemistry 43:1754-1762. [DOI] [PubMed] [Google Scholar]
- 38.Ramirez-Aguilar, K. A., N. A. Low-Nam, and R. D. Kuchta. 2002. Key role of template sequence for primer synthesis by the herpes simplex virus 1 helicase-primase. Biochemistry 41:14569-14579. [DOI] [PubMed] [Google Scholar]
- 39.Salinas, F., and S. J. Benkovic. 2000. Characterization of bacteriophage T4-coordinated leading- and lagging-strand synthesis on a minicircle substrate. Proc. Natl. Acad. Sci. U. S. A. 97:7196-7201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Severini, A., D. G. Scraba, and D. L. J. Tyrrell. 1996. Branched structures in the intracellular DNA of herpes simplex virus type 1. J. Virol. 70:3169-3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Severson, J. L., and S. K. Tyring. 1999. Viral disease update. Curr. Probl. Dermatol. 11:43-70. [Google Scholar]
- 42.Sherman, G., J. Gottlieb, and M. D. Challberg. 1992. The UL8 subunit of the herpes simplex virus helicase-primase complex is required for efficient primer utilization. J. Virol. 66:4884-4892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Skaliter, R., A. M. Makhov, J. D. Griffith, and I. R. Lehman. 1996. Rolling circle DNA replication by extracts of herpes simplex virus type 1-infected human cells. J. Virol. 70:1132-1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Song, L. P., M. Chaudhuri, C. W. Knopf, and D. S. Parris. 2004. Contribution of the 3′- to 5′-exonuclease activity of herpes simplex virus type 1 DNA polymerase to the fidelity of DNA synthesis. J. Biol. Chem. 279:18535-18543. [DOI] [PubMed] [Google Scholar]
- 45.Strang, B. L., and N. D. Stow. 2005. Circularization of the herpes simplex virus type 1 genome upon lytic infection. J. Virol. 79:12487-12494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tanguy Le Gac, N., G. Villani, J. S. Hoffmann, and P. E. Boehmer. 1996. The UL8 subunit of the herpes simplex virus type-1 DNA helicase-primase optimizes utilization of DNA templates covered by the homologous single-strand DNA-binding protein ICP8. J. Biol. Chem. 271:21645-21651. [DOI] [PubMed] [Google Scholar]
- 47.Taylor, T. J., and D. M. Knipe. 2004. Proteomics of herpes simplex virus replication compartments: association of cellular DNA replication, repair, recombination, and chromatin remodeling proteins with ICP8. J. Virol. 78:5856-5866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tenney, D. J., W. W. Hurlburt, P. A. Micheletti, M. Bifano, and R. K. Hamatake. 1994. The UL8 component of the herpes simplex virus helicase-primase complex stimulates primer synthesis by a subassembly of the UL5 and UL52 components J. Biol. Chem. 269:5030-5035. [PubMed] [Google Scholar]
- 49.Weisshart, K., C. S. Chow, and D. M. Coen. 1999. Herpes simplex virus processivity factor UL42 imparts increased DNA-binding specificity to the viral DNA polymerase and decreased dissociation from primer-template without reducing the elongation rate. J. Virol. 73:55-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wilcock, D., and D. P. Lane. 1991. Localization of p53, retinoblastoma and host replication proteins at sites of viral replication in herpes-infected cells. Nature 349:429-431. [DOI] [PubMed] [Google Scholar]
- 51.Wu, C. A., E. L. Zechner, J. A. Reems, C. S. McHenry, and K. J. Marians. 1992. Coordinated leading-strand and lagging-strand synthesis at the Escherichia coli DNA replication fork V. Primase action regulates the cycle of Okazaki fragment synthesis. J. Biol. Chem. 267:4074-4083. [PubMed] [Google Scholar]
- 52.Yang, J., S. W. Nelson, and S. J. Benkovic. 2006. The control mechanism for lagging strand polymerase recycling during bacteriophage T4 DNA replication. Mol. Cell 21:153-164. [DOI] [PubMed] [Google Scholar]
- 53.Zhu, Y. L., K. S. Trego, L. P. Song, and D. S. Parris. 2003. 3′ to 5′ Exonuclease activity of herpes simplex virus type 1 DNA polymerase modulates its strand displacement activity. J. Virol. 77:10147-10153. [DOI] [PMC free article] [PubMed] [Google Scholar]