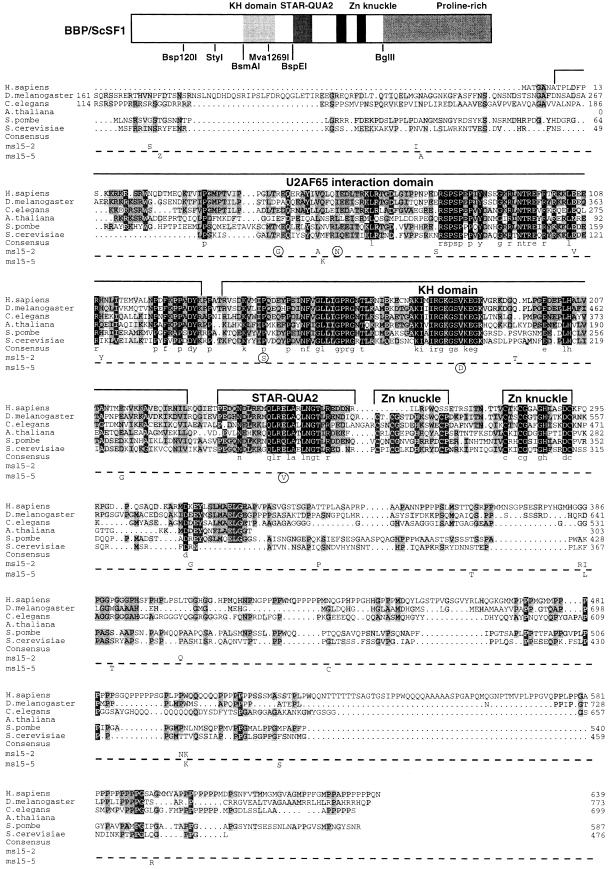
Fig. 1. Amino acid sequence alignment of SF1 orthologs and position of mutations in msl5-2 and msl5-5. Putative homologs of SF1 from Homo sapiens, Drosophila melanogaster, Caenorhabditis elegans, Arabidopsis thaliana (partial sequence), Schizosaccharomyces pombe and Saccharomyces cerevisae were aligned using the program ClustalX (Thompson et al., 1997) and manual refinement. Amino acids are shaded according to the degree of conservation. Functional domains and structural motifs are indicated above the alignment. The U2AF65 interaction domain is drawn according to the results published for HsSF1 (Rain et al., 1998). The positions of point mutations in the cs mutant msl5-5 (below the dashed line) and in the ts mutant msl5-2 (above the dashed line) are indicated below the S.cerevisiae SF1 sequence. Mutations that were sufficient and necessary to retain a growth defect are encircled (compare with Figure 2). Sequences have the following DDBJ/EMBL/GenBank accession Nos: AtSF1, AB023044; CeSF1, AJ243905; DmSF1, AJ243904; HsSF1, Y08765; ScSF1, U53877; SpSF1, SPTREMBL O74555. The partial protein sequence of A.thaliana SF1 was generated by comparison of the genomic sequence with a profile (derived from an alignment of the known SF1 orthologs with the program PROFILEWEIGHT; Thompson et al., 1994) using the program PairWise (Birney et al., 1996). Restriction enzymes used for domain swapping between mutants and wild-type ORF are shown below the cartoon, which depicts the structural organization of BBP/ScSF1.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.