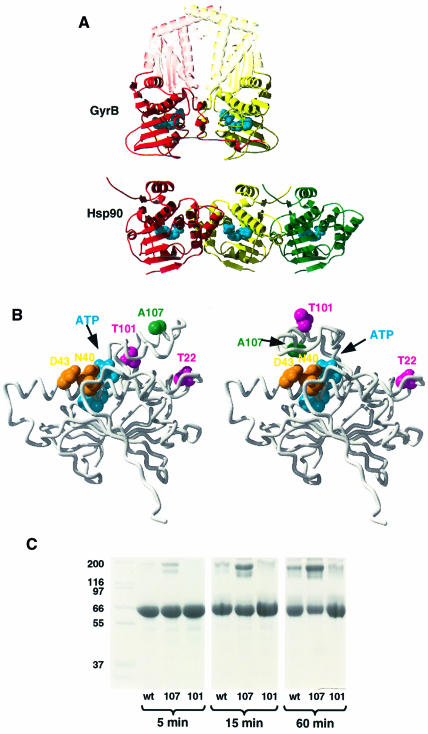
Fig. 7. Hsp90 N-terminal dimer interface and lid closure. (A) Comparison of GyrB 40 kDa N-terminal fragment dimer (top) with alternative dimers in the yeast Hsp90 N-domain crystal lattice (bottom). The relative orientation of the leftmost pair (red–yellow) of Hsp90 N-domains more closely resembles that of the N-domains in GyrB than does the rightmost pair (yellow–green), originally suggested. The conformations of the ‘lid’ segments and the N-terminal strands differ between GyrB and Hsp90. AMP-PNP bound to GyrB and ADP bound to Hsp90 are shown as blue CPK models. (B) Comparison of the open conformation of the ‘lid’ segment observed in the crystal structure of yeast Hsp90 N-domain (left) with the modelled structure of the closed conformation (right). Ala107, which is exposed in the open conformation, comes close to a hydrophilic patch provided by residues Ser39, Asp40 and Asp43 in the closed conformation. Its mutation to asparagine, which is expected to stabilize this conformation, produces a dramatic increase in ATPase activity (see text). The positions of Thr22 and Thr101 are also indicated. (C) Polyacrylamide gel showing cross-linking as a function of incubation time for the C-terminal truncation mutant NΔ0C551 (wt), the A107N-NΔ0C551 double mutant (107) and the T101I-NΔ0C551 double mutant (101) cross-linked with a 15-fold molar excess of DMS, in the presence of AMP-PNP.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.