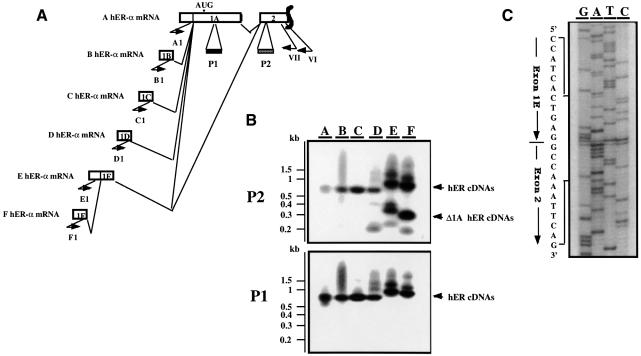
Fig. 2. Exon 1E is alternatively spliced to exon 1A or exon 2. (A) Schematic representation of the RT–PCR experiment designed to identify Δ1A hER-α mRNAs. Open boxes indicate the unique (1A–1F) and the two first common (1A, 2) exons encoding each hER-α mRNA variant. Approximate locations of primers are shown by short arrows. Primer VI, located in exon 2, was used to prime hER-α cDNA synthesis by reverse transcriptase. Primers A1–F1, which are specific for each hER-α cDNA 5′ region, were used in a round of PCR amplification with primer VII, which is nested to primer VI in exon 2. The oligonucleotide probes P1 and P2 from exon 1A and 2, respectively, were used to confirm the specificity of the PCR products as well as the exon 1A deletion for some hER-α transcripts. (B) The hER-α cDNA variants were amplified as described above, using total RNA from MCF7. PCR products were electrophoresed through an agarose gel and transferred by Southern blotting to a membrane, which was then hybridized with the oligonucleotide probes P1 and P2 as described in Materials and methods. Positions of migration of the molecular size markers are shown on the left side of the figure. (C) The sequence of the PCR products from lane E or F (B) that did not hybridize to the oligonucleotide probe P1 but hybridized to P2 probe revealed that they contain the donor site of exon 1E joined to the acceptor site of exon 2.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.