Abstract
In a 2-year prospective study of tuberculosis (TB) patients in China, the prevalences of non-Beijing strains of Mycobacterium tuberculosis varied between Shandong Province (20.6%), Shanghai (27.6%), and Sichuan Province (45.9%) (P < 0.005). These differences may be due to factors such as human migration, transmission, or diversification and adaptation of the mycobacteria to different hosts.
In 1995, van Soolingen reported that 86% of Mycobacterium tuberculosis strains isolated from tuberculosis patients in the Beijing area of China had highly similar multiband IS6110 restriction fragment length polymorphism (RFLP) patterns and characteristic spoligotyping patterns. These strains were called Beijing family strains (25). Later, the multidrug-resistant “W” strain of M. tuberculosis that caused an outbreak in New York City in 1991 (4) was recognized as a member of the Beijing family strains (14). Since then, Beijing family strains have been isolated from many populations throughout the world and have been reported to have a high prevalence (8, 20), high virulence (17, 24), and a high probability of drug resistance (2, 12).
Although Beijing family strains were assumed to be the predominant strains causing active tuberculosis in China, different sublineages of non-Beijing strains have been detected in the countries and regions adjacent to mainland China (1, 3, 22, 23). In Taiwan, strains in the Haarlam (H) sublineage, East African-Indian (EAI) and EAI-like sublineages, and Latin American-Mediterranean (LAM) sublineage are also prevalent (7, 9). Three of the most hypervirulent clinical strains isolated in a study in Shanghai and Hong Kong belonged to families of non-Beijing strains (26). Furthermore, the prevalence of Beijing strains varied from 25% to 91.7% in different studies from different areas in mainland China (5, 16). Taken together, these findings suggest that non-Beijing strains may also contribute to the high tuberculosis burden in Asia.
To determine the prevalence and transmission potential of non-Beijing family strains in mainland China, we performed a population-based prospective study in three geographic areas. Fei County and Yan Zhou City are in Shandong Province, located in the central eastern part of China. Songjiang and Chongming are two districts in the municipality of Shanghai, south of Shandong Province. Shuangliu County is in Sichuan Province in the southwestern region of China. We collected 1,004 M. tuberculosis clinical isolates from 988 culture-positive pulmonary tuberculosis patients from 1 December 2006 to 31 December 2008. We selected the initial isolate from each patient for a total of 988 isolates that were analyzed in this study. M. tuberculosis strains from sputum samples were cultured with Lowenstein-Jensen (L-J) medium (18). Of the total of 988 isolates in the present study, 381 (38.6%) were from Shanghai, 315 (31.9%) were from Shandong Province, and 292 (29.5%) were from Sichuan Province. For each of the clinical isolates, genomic DNA was extracted from the mycobacterial culture by the protocol described by Shen et al. (21). Three methods were used to genotype the strains. First, all strains were screened by using a deletion-targeted multiplex PCR (DTM-PCR) method to rapidly identify Beijing family strains, followed by 7-locus and 16-locus variable number of tandem repeats (VNTR) genotyping methods (6, 27). The non-Beijing strains that were identified by DTM-PCR were also spoligotyped (3, 11).
Among the 988 clinical isolates, 304 (30.8%) had non-Beijing strains of M. tuberculosis. Of the 988 patients, demographic information was missing from 4 of the tuberculosis patients. We used the nonparametric Wilcoxon rank sum test and the chi-square test of proportions to analyze the data. The median age of tuberculosis patients with a non-Beijing family strain (45.7 years; range, 18 to 87 years) was not significantly different from the median age of tuberculosis patients with a Beijing family strain (48.9 years; range, 15 to 98 years) (P = 0.66). The proportion of males among the tuberculosis patients with a non-Beijing family strain of M. tuberculosis (73.8%) did not differ from the proportion of males among the tuberculosis patients with a Beijing family strain (74.4%) (P = 0.86).
The percentage of non-Beijing strains varied in different geographic areas: 45.9% of the strains isolated from Sichuan were non-Beijing strains, while only 20.6% of the strains in Shandong Province were non-Beijing strains (P < 0.0005) (Table 1). The proportion of non-Beijing family strains in Sichuan Province was significantly higher than those in Shandong Province (P < 0.0005) and Shanghai (P < 0.0005). The proportion of non-Beijing strains in Shanghai was also significantly higher than that in Shandong Province (P = 0.034). Therefore, the prevalences of non-Beijing strains were different across the three study areas in mainland China.
TABLE 1.
Frequency and percentage of Beijing and non-Beijing family strains of M. tuberculosis in different areas of mainland China, 1 December 2006 to 31 December 2008
| Sublineage | Frequency or percentage of strains in: |
P value | |||||
|---|---|---|---|---|---|---|---|
| Shandong |
Shanghai |
Sichuan |
|||||
| n | % | n | % | n | % | ||
| Total | 315 | 100 | 381 | 100 | 292 | 100 | |
| Beijing family strains | 250 | 79.4 | 276 | 72.4 | 158 | 54.8 | Reference |
| Non-Beijing family strains | 65 | 20.6 | 105 | 27.6 | 134 | 45.2 | <0.0005 |
| T1 | 18 | 5.7 | 34 | 8.9 | 55 | 18.5 | |
| T2 | 25 | 7.9 | 26 | 6.8 | 26 | 8.5 | |
| H3 | 1 | 0.3 | 3 | 0.8 | 9 | 3.1 | |
| Othersa | 5 | 1.6 | 10 | 2.6 | 9 | 3.1 | |
| Unclassified | 16 | 5.1 | 32 | 8.4 | 35 | 12.0 | |
The category “Others” includes sublineages H1, H4, LAM10_CAM, LAM9, MANU2, S, T1-T2, T2-T3, T3, T5, U (likely T3), and X.
Based on the samples isolated in a nationwide random survey in 2000, the data from Li et al. showed that the difference in the prevalence of Beijing strains by region was of borderline significance (P = 0.06) (15). In our study, Beijing family strains were more prevalent in the central eastern part of China (Fei County and Yan Zhou County in Shandong Province), areas which have been inhabited for many centuries and have expansive, wide-reaching transportation systems. Throughout Chinese history, large numbers of people have migrated to and from the central eastern regions, likely disseminating the Beijing family strains. In contrast, Sichuan Province in southwestern China has a rather rugged geographical environment, its transportation systems were developed later, and non-Beijing strains are more prevalent in this region.
The spoligotypes of the 304 non-Beijing strains were compared with the data available in the fourth version of the international spoligotype database, SpolDB4 (http://www.pasteur-guadeloupe.fr/tb/spoldb4/spoldb4.pdf; accessed 31 January 2010). Altogether, we detected 113 different spoligotypes among the 304 isolates: 75 isolates (24.7%) displayed unique spoligotypes, and 229 (75.3%) had 1 of 38 spoligotypes. The discriminatory power (according to the Hunter-Gaston index [HGI]) of the spoligotyping method among non-Beijing strains in our study areas was 0.92 (13). Spoligotyping has a rather low discriminatory power among Beijing family strains, but when combined with DTM-PCR, it still provides a useful, rapid method to genotype non-Beijing strains in China.
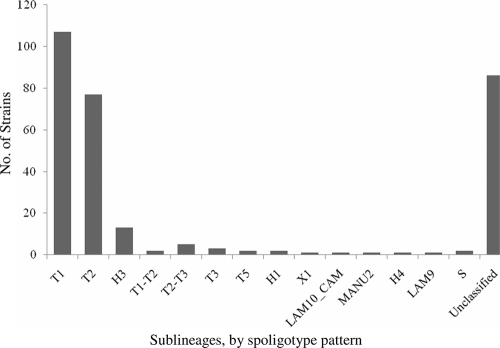
We classified the spoligotypes from 219 isolates (72.0%; 219/304) into 53 shared international types (STs). The remaining 85 strains had 61 different spoligotypes that were not identified in the database. Of the 53 spoligotypes that were in SpolDB4, the most prevalent STs were ST52 in the T2 sublineage (27.9%; 61/219), followed by ST53 of the T1 sublineage (26.0%; 57/219), and ST50 of the Haarlem3 (H3) sublineage (4.6%, 10/219) (Table 2). The T family was the most prevalent sublineage, present in 196 (89.5%, 196/219) isolates. Among the strains in the T family, 107 belonged to the T1 sublineage, and 77 belonged to the T2 sublineage. The second largest family of non-Beijing strains was the H sublineage, with 16 isolates (7.3%; 16/219) (Table 2 and Fig. 1).
TABLE 2.
Spoligotypes of 219 isolates of M. tuberculosis with a shared international type number in the SpolDB4 database for mainland China from 1 December 2006 to 31 December 2008
| Spoligotype | Sublineage | STa | No. of isolates |
|---|---|---|---|
| 777777774020771 | H1 | 47 | 2 |
| 777777770020771 | H3 | 742 | 2 |
| 777777777520771 | H3 | 746 | 1 |
| 777777777720771 | H3 | 50 | 10 |
| 577777777420771 | H4 | 127 | 1 |
| 777777743760731 | LAM10_CAM | 403 | 1 |
| 737777607760771 | LAM9 | 388 | 1 |
| 777777777763771 | MANU2 | 54 | 1 |
| 576377777760771 | S | 1211 | 2 |
| 377777777760771 | T1 | 7 | 2 |
| 477777777760771 | T1 | 804 | 1 |
| 577777777760771 | T1 | 334 | 6 |
| 637777777760771 | T1 | 285 | 1 |
| 677777777760771 | T1 | 196 | 2 |
| 737777777760771 | T1 | 205 | 1 |
| 776037777760771 | T1 | 210 | 1 |
| 776777777760771 | T1 | 1129 | 1 |
| 777577777760771 | T1 | 917 | 1 |
| 777617777760771 | T1 | 1214 | 3 |
| 777640007760771 | T1 | 249 | 1 |
| 777677777760771 | T1 | 498 | 1 |
| 777717777760771 | T1 | 131 | 7 |
| 777740007760771 | T1 | 803 | 1 |
| 777743777760771 | T1 | 913 | 1 |
| 777757777760771 | T1 | 393 | 2 |
| 777763777760771 | T1 | 732 | 1 |
| 777775777760771 | T1 | 281 | 1 |
| 777777403760771 | T1 | 1688 | 2 |
| 777777407760771 | T1 | 1800 | 1 |
| 777777707760771 | T1 | 535 | 1 |
| 777777737760771 | T1 | 86 | 3 |
| 777777774760771 | T1 | 353 | 1 |
| 777777776760771 | T1 | 1626 | 1 |
| 777777777760631 | T1 | 888 | 2 |
| 777777777760700 | T1 | 51 | 3 |
| 777777777760740 | T1 | 1583 | 1 |
| 777777777760761 | T1 | 278 | 1 |
| 777777777760771 | T1 | 53 | 57 |
| 777777777760711 | T1-T2 | 78 | 2 |
| 577777777760731 | T2 | 1302 | 3 |
| 737777777760731 | T2 | 848 | 1 |
| 747777777760731 | T2 | 712 | 2 |
| 757777777760731 | T2 | 153 | 5 |
| 777767777760771 | T2 | 118 | 1 |
| 777777377760731 | T2 | 1077 | 1 |
| 777777707760731 | T2 | 1890 | 2 |
| 777777770760731 | T2 | 942 | 1 |
| 777777777760731 | T2 | 52 | 61 |
| 777737777760731 | T2-T3 | 73 | 5 |
| 777737777760771 | T3 | 37 | 3 |
| 777777757760771 | T5 | 44 | 2 |
| 777737770000000 | U (likely T3) | 56 | 1 |
| 777776777760771 | X1 | 119 | 1 |
ST, shared international type number.
FIG. 1.
Number of isolates with non-Beijing family strains of M. tuberculosis in three different geographical areas, by sublineages, from 1 December 2006 to 31 December 2008. The major sublineages of non-Beijing strains detected in the study were T1 and T2, which can be found on most continents in the world. “Unclassified” includes a group of strains with spoligotyping patterns that were not classified in SpolDB4.
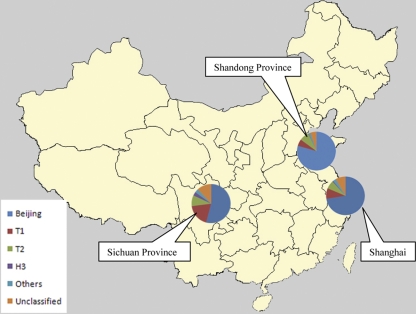
The same sublineages of non-Beijing family strains were prevalent in our three different study areas (Fig. 2), but the distribution of sublineages of M. tuberculosis by spoligotypes varies throughout the world (11). For example, the Beijing family strains are prevalent in the Far East-Asia (19), and the EAI family was predominant in Southeast Asian countries (10, 19). H sublineages of M. tuberculosis have been found in Europe, Central America, and the Caribbean; LAM sublineages are prevalent in South America, Africa, the Mediterranean basin, and the Caribbean region (3). In the countries adjacent to China, EAI family, Central Asia strains (CAS) were reported in neighboring Vietnam and Thailand, Pakistan, and India (1, 3, 22, 23). The predominant non-Beijing strains in our study were in the T family, which remains ill defined and has been reported from all continents (3). Further studies are needed to improve our knowledge of the factors and selective pressures that determine the prevalence of these strains in China.
FIG. 2.
Map of China showing the percentage of different sublineages of M. tuberculosis in three different geographical areas from 1 December 2006 to 31 December 2008. The category “Others” includes sublineages H1, H4, LAM10_CAM, LAM9, MANU2, S, T1-T2, T2-T3, T3, T5, U (likely T3), and X.
To determine whether recent transmission of M. tuberculosis was likely in our study population, we compared the genotypes obtained by VNTR methods. For the strains with identical VNTR-7 patterns, VNTR-16 was performed. Strains from tuberculosis patients in the same study site (Shanghai, Shandong Province, or Sichuan Province) with identical VNTR-16 and spoligotyping patterns were defined as clustered strains (see Table S1 in the supplemental material). Among 304 non-Beijing strains, we found 15 VNTR clusters with a total of 39 strains. Among the 15 clusters, 3 clusters with 9 strains belonged to the T1 sublineage, 6 clusters with 17 strains belonged to the T2 sublineage, 1 cluster with two strains belonged to the T2-T3 sublineage, and five clusters with 11 strains had a previously unclassified spoligotype pattern. Strains in the T1 sublineage were less likely to be clustered than Beijing family strains (P = 0.0001) (Table 3).
TABLE 3.
Sublineages of clustered and nonclustered clinical isolates of M. tuberculosis, based on spoligotyping and VNTR-7 typing patterns
| Spoligotyping lineage or sublineagea | VNTRb result from: |
P value | |||
|---|---|---|---|---|---|
| Clustered strains |
Nonclustered strains |
||||
| n | % | n | % | ||
| Beijing family strains | 176 | 25.7 | 508 | 74.3 | Reference |
| Non-Beijing family strains | |||||
| T1 | 9 | 8.4 | 98 | 91.6 | 0.0001 |
| T2 | 17 | 22.1 | 60 | 77.9 | 0.485 |
| T2-T3 | 2 | 40.0 | 3 | 60.0 | 0.468 |
| Unclassified | 11 | 12.9 | 74 | 87.1 | 0.010 |
Strains in the T1 sublineage were less likely to be clustered than Beijing family strains (P < 0.05). “Unclassified” includes the different spoligotyping patterns that were not identified in the SpolDB4 database.
VNTR, variable number of tandem repeats.
In conclusion, despite the high prevalence of Beijing family strains in mainland China, many non-Beijing strains were also isolated from tuberculosis patients. Although the sublineages of non-Beijing strains were similar between different populations, the prevalence of non-Beijing sublineages of strains varied across different geographical areas. The most prevalent non-Beijing family strains in our study population belonged to the T family, which has been reported from all continents. Compared with Beijing family strains, strains in the T1 sublineage were less likely to be clustered.
Supplementary Material
Acknowledgments
We thank Sebastien Gagneux for valuable comments on the manuscript.
This work was supported by the Key Project of Chinese National Programs (2009ZX10003-017) and National Institutes of Health (NIH) grant D43 TW007887. This work is also part of the TB-VIR Network (Collaborative Project) supported by the European Commission under the Health Cooperation Work Programme of the 7th Framework Programme (grant agreement 200973).
Footnotes
Published ahead of print on 10 November 2010.
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1.Arora, J., U. B. Singh, N. Suresh, T. Rana, C. Porwal, A. Kaushik, and J. N. Pande. 2009. Characterization of predominant Mycobacterium tuberculosis strains from different subpopulations of India. Infect. Genet. Evol. 9:832-839. [DOI] [PubMed] [Google Scholar]
- 2.Borrell, S., and S. Gagneux. 2009. Infectiousness, reproductive fitness and evolution of drug-resistant Mycobacterium tuberculosis. Int. J. Tuberc. Lung Dis. 13:1456-1466. [PubMed] [Google Scholar]
- 3.Brudey, K., J. R. Driscoll, L. Rigouts, W. M. Prodinger, A. Gori, S. A. Al-Hajoj, C. Allix, L. Aristimuno, J. Arora, V. Baumanis, L. Binder, P. Cafrune, A. Cataldi, S. Cheong, R. Diel, C. Ellermeier, J. T. Evans, M. Fauville-Dufaux, S. Ferdinand, D. Garcia de Viedma, C. Garzelli, L. Gazzola, H. M. Gomes, M. C. Guttierez, P. M. Hawkey, P. D. van Helden, G. V. Kadival, B. N. Kreiswirth, K. Kremer, M. Kubin, S. P. Kulkarni, B. Liens, T. Lillebaek, M. L. Ho, C. Martin, I. Mokrousov, O. Narvskaia, Y. F. Ngeow, L. Naumann, S. Niemann, I. Parwati, Z. Rahim, V. Rasolofo-Razanamparany, T. Rasolonavalona, M. L. Rossetti, S. Rusch-Gerdes, A. Sajduda, S. Samper, I. G. Shemyakin, U. B. Singh, A. Somoskovi, R. A. Skuce, D. van Soolingen, E. M. Streicher, P. N. Suffys, E. Tortoli, T. Tracevska, V. Vincent, T. C. Victor, R. M. Warren, S. F. Yap, K. Zaman, F. Portaels, N. Rastogi, and C. Sola. 2006. Mycobacterium tuberculosis complex genetic diversity: mining the Fourth International Spoligotyping Database (SpolDB4) for classification, population genetics and epidemiology. BMC Microbiol. 6:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention. 1993. Outbreak of multidrug-resistant tuberculosis at a hospital—New York City, 1991. MMWR Morb. Mortal. Wkly. Rep. 42:427, 433-434. [PubMed] [Google Scholar]
- 5.Chai, L. Q., W. M. Li, L. Li, Z. J. Dai, D. P. Bai, L. Zhang, S. F. Shao, Q. Wu, W. Lu, Z. G. Sun, and C. Y. Li. 2007. Study on the genotype of Mycobacterium tuberculosis isolates from hospitals in Tianjin. Zhonghua Liu Xing Bing Xue Za Zhi. 28:785-788. (In Chinese.) [PubMed] [Google Scholar]
- 6.Chen, J., A. G. Tsolaki, X. Shen, X. Jiang, J. Mei, and Q. Gao. 2007. Deletion-targeted multiplex PCR (DTM-PCR) for identification of Beijing/W genotypes of Mycobacterium tuberculosis. Tuberculosis (Edinb.) 87:446-449. [DOI] [PubMed] [Google Scholar]
- 7.Chuang, P. C., H. Liu, C. Sola, Y. M. Chen, and R. Jou. 2008. Spoligotypes of Mycobacterium tuberculosis isolates of a high tuberculosis burden aboriginal township in Taiwan. Infect. Genet. Evol. 8:553-557. [DOI] [PubMed] [Google Scholar]
- 8.Cowley, D., D. Govender, B. February, M. Wolfe, L. Steyn, J. Evans, R. J. Wilkinson, and M. P. Nicol. 2008. Recent and rapid emergence of W-Beijing strains of Mycobacterium tuberculosis in Cape Town, South Africa. Clin. Infect. Dis. 47:1252-1259. [DOI] [PubMed] [Google Scholar]
- 9.Dou, H. Y., F. C. Tseng, C. W. Lin, J. R. Chang, J. R. Sun, W. S. Tsai, S. Y. Lee, I. J. Su, and J. J. Lu. 2008. Molecular epidemiology and evolutionary genetics of Mycobacterium tuberculosis in Taipei. BMC Infect. Dis. 8:170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Douglas, J. T., L. Qian, J. C. Montoya, J. M. Musser, J. D. Van Embden, D. Van Soolingen, and K. Kremer. 2003. Characterization of the Manila family of Mycobacterium tuberculosis. J. Clin. Microbiol. 41:2723-2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Driscoll, J. R. 2009. Spoligotyping for molecular epidemiology of the Mycobacterium tuberculosis complex. Methods Mol. Biol. 551:117-128. [DOI] [PubMed] [Google Scholar]
- 12.Glynn, J. R., J. Whiteley, P. J. Bifani, K. Kremer, and D. van Soolingen. 2002. Worldwide occurrence of Beijing/W strains of Mycobacterium tuberculosis: a systematic review. Emerg. Infect. Dis. 8:843-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hunter, P. R., and M. A. Gaston. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J. Clin. Microbiol. 26:2465-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kurepina, N. E., S. Sreevatsan, B. B. Plikaytis, P. J. Bifani, N. D. Connell, R. J. Donnelly, D. van Sooligen, J. M. Musser, and B. N. Kreiswirth. 1998. Characterization of the phylogenetic distribution and chromosomal insertion sites of five IS6110 elements in Mycobacterium tuberculosis: non-random integration in the dnaA-dnaN region. Tuber. Lung Dis. 79:31-42. [DOI] [PubMed] [Google Scholar]
- 15.Li, W. M., S. M. Wang, C. Y. Li, Y. H. Liu, G. M. Shen, X. X. Zhang, T. G. Niu, Q. Gao, D. van Soolingen, K. Kremer, and H. J. Duanmu. 2005. Molecular epidemiology of Mycobacterium tuberculosis in China: a nationwide random survey in 2000. Int. J. Tuberc. Lung Dis. 9:1314-1319. [PubMed] [Google Scholar]
- 16.Li, W. M., S. M. Wang, X. Y. Pei, Z. Q. Liu, Q. Zhong, M. Qian, B. Zhao, H. J. Duanmu. 2003. DNA fingerprinting of Mycobacterium tuberculosis strains from Beijing, Guangdong and Ningxia. Zhonghua Liu Xing Bing Xue Za Zhi 24:381-417. [PubMed] [Google Scholar]
- 17.Manca, C., L. Tsenova, S. Freeman, A. K. Barczak, M. Tovey, P. J. Murray, C. Barry, and G. Kaplan. 2005. Hypervirulent M. tuberculosis W/Beijing strains upregulate type I IFNs and increase expression of negative regulators of the Jak-Stat pathway. J. Interferon Cytokine Res. 25:694-701. [DOI] [PubMed] [Google Scholar]
- 18.Paniker, C. K. J. 2005. Ananthanarayan and Paniker's textbook of microbiology, 7th ed. Mycobacterium. I. Tuberculosis. Orient Longman Private, Ltd., Hyderabad, India.
- 19.Phyu, S., R. Jureen, T. Ti, U. R. Dahle, and H. M. Grewal. 2003. Heterogeneity of Mycobacterium tuberculosis isolates in Yangon, Myanmar. J. Clin. Microbiol. 41:4907-4908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qian, L., J. D. Van Embden, A. G. Van Der Zanden, E. F. Weltevreden, H. Duanmu, and J. T. Douglas. 1999. Retrospective analysis of the Beijing family of Mycobacterium tuberculosis in preserved lung tissues. J. Clin. Microbiol. 37:471-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shen, G., Z. Xue, X. Shen, B. Sun, X. Gui, M. Shen, J. Mei, and Q. Gao. 2006. Recurrent tuberculosis and exogenous reinfection, Shanghai, China. Emerg. Infect. Dis. 12:1776-1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Singh, U. B., J. Arora, N. Suresh, H. Pant, T. Rana, C. Sola, N. Rastogi, and J. N. Pande. 2007. Genetic biodiversity of Mycobacterium tuberculosis isolates from patients with pulmonary tuberculosis in India. Infect. Genet. Evol. 7:441-448. [DOI] [PubMed] [Google Scholar]
- 23.Tanveer, M., Z. Hasan, A. R. Siddiqui, A. Ali, A. Kanji, S. Ghebremicheal, and R. Hasan. 2008. Genotyping and drug resistance patterns of M. tuberculosis strains in Pakistan. BMC Infect. Dis. 8:171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsenova, L., E. Ellison, R. Harbacheuski, A. L. Moreira, N. Kurepina, M. B. Reed, B. Mathema, C. E. Barry III, and G. Kaplan. 2005. Virulence of selected Mycobacterium tuberculosis clinical isolates in the rabbit model of meningitis is dependent on phenolic glycolipid produced by the bacilli. J. Infect. Dis. 192:98-106. [DOI] [PubMed] [Google Scholar]
- 25.van Soolingen, D., L. Qian, P. E. de Haas, J. T. Douglas, H. Traore, F. Portaels, H. Z. Qing, D. Enkhsaikan, P. Nymadawa, and J. D. van Embden. 1995. Predominance of a single genotype of Mycobacterium tuberculosis in countries of east Asia. J. Clin. Microbiol. 33:3234-3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wong, K. C., W. M. Leong, H. K. Law, K. F. Ip, J. T. Lam, K. Y. Yuen, P. L. Ho, W. S. Tse, X. H. Weng, W. H. Zhang, S. Chen, and W. C. Yam. 2007. Molecular characterization of clinical isolates of Mycobacterium tuberculosis and their association with phenotypic virulence in human macrophages. Clin. Vaccine Immunol. 14:1279-1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang, L., J. Chen, X. Shen, X. Gui, J. Mei, K. DeRiemer, and Q. Gao. 2008. Highly polymorphic variable-number tandem repeats loci for differentiating Beijing genotype strains of Mycobacterium tuberculosis in Shanghai, China. FEMS Microbiol. Lett. 282:22-31. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.