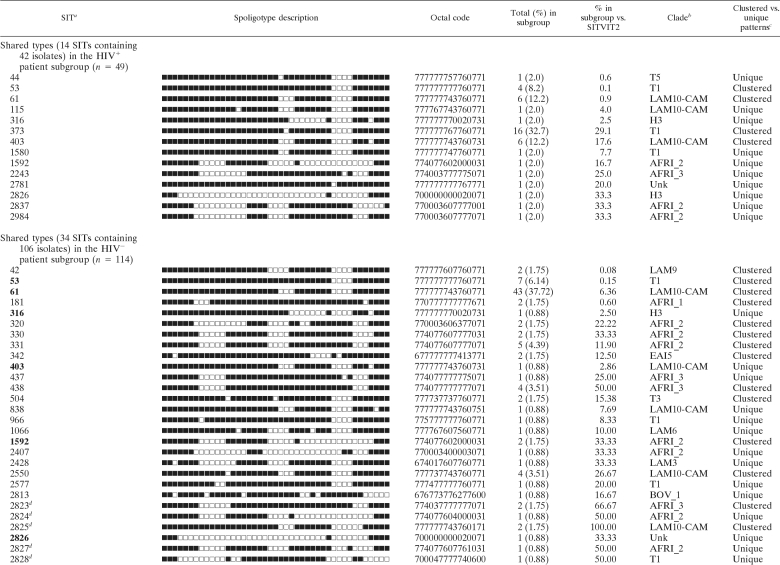
TABLE 2.
Spoligotyping shared types from HIV+ and HIV− patient subgroups in Ibadan, Nigeria
a Unique strains matching a preexisting pattern in the SITVIT2 database are classified as SITs, whereas in cases where there was no match, they are designated “orphan” (Table 1). SITs in the HIV− subgroup that were simultaneously present in the HIV+ subgroup are in boldface.
b Clade designations according to SITVIT2 using revised SpolDB4 rules. Unk, unknown patterns within any of the major clades described.
c Clustered strains correspond to a similar spoligotype pattern shared by 2 or more strains within this study as opposed to unique strains harboring a spoligotype pattern that does not match another strain from this study.
dNewly created shared type (none among 14 SITs from HIV+ patients as opposed to 11 new SITs observed among 34 SITs from HIV− patients). In the latter subgroup, these new SITs were created due to 2 or more strains belonging to an identical new pattern within this study (SIT2825, n = 2; SIT2829, n = 2; SIT2831, n = 3), a unique strain from this study matching another orphan in the database (SIT2824 matched an orphan from metropolitan France; SIT2830, SIT2832, and SIT2834 each matched an orphan from the United States), or 2 or more strains from this study that matched an existing orphan from Nigeria (SIT2823, n = 2; SIT2827, n = 1; SIT2828, n = 1; SIT2835, n = 3).