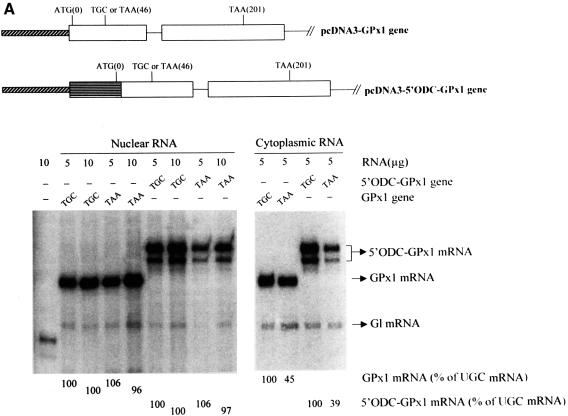
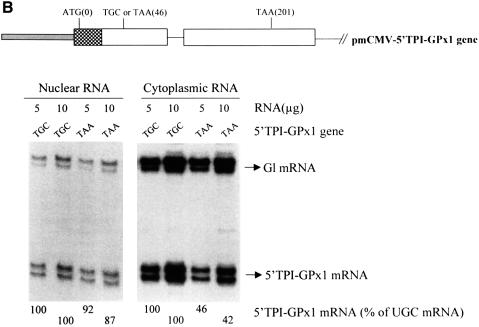
Fig. 4. Neither the extent nor cellular site of GPx1 NMD is affected by substituting the GPx1 5′-UTR and beginning of the coding region within the ODC translational enhancer and beginning of the CAT gene or the corresponding regions of the TPI gene. (A) Structures of the pcDNA3-GPx1 and pcDNA3-5′ODC-GPx1 genes. The diagonally striped bar represents the CMV promoter of pcDNA3 and the horizontally striped exonic region represents part of the 5′-UTR of ODC and the first four codons of the CAT gene. Cos cells were transiently transfected with the specified GPx1 test allele and the Gl reference allele. Nuclear and cytoplasmic RNA was purified (Zhang et al., 1998b), and 5 or 10 µg were analyzed by RNase mapping using uniformly 32P-labeled antisense RNAs (see Materials and methods). Cytoplasmic RNA was exposed to X-ray film for a shorter time than nuclear RNA. The analysis of untransfected cells demonstrated that RNase-resistant 32P measured either test or reference RNAs. The finding that 10 µg of RNA protected ∼2-fold more 32P than 5 µg of RNA demonstrated that the analysis was quantitative. The level of each GPx1 and 5′ODC-GPx1 mRNA was normalized to the level of Gl mRNA and is presented as a percentage of the level of the corresponding UGC-containing mRNA, which was defined as 100. Results are the average of three independently performed experiments and did not differ by >7%. (B) Structure of the 5′TPI-GPx1 gene. The gray bar/attached checkered exonic region represent, respectively, the mCMV promoter/5′-UTR and first 22 codons of the TPI gene. RNase mapping and quantitation of the level of 5′TPI-GPx1 mRNA were as described for (A). For both (A) and (B), antisense RNAs that appear as RNase-resistant doublets reflect incomplete or excessive RNase digestion rather than structural heterogeneity of the mRNA under analysis.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.