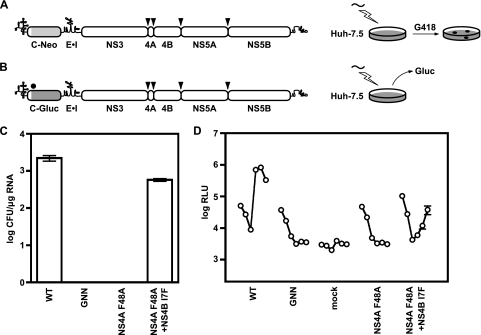
FIG. 3.
NS4B I7F partially suppresses the replication defect of subgenomic replicons containing the NS4A F48A mutation. (A) The pYSGR-JFH construct used in the present study and the workflow to identify revertants or suppressor mutants. HCV polyprotein processing sites are annotated as in Fig. 1. (B) pYSGR-JFH/GLuc reporter construct used in this study and workflow of our reporter virus assay. RNA was electroporated into Huh-7.5 cells, and culture medium was harvested at various times posttransfection. Secreted GLuc activity was assayed as detailed in Materials and Methods. (C) Stable replication phenotypes of NS4A F48A and its suppressor NS4B I7F. Colony-forming activity was measured for NS4A F48A and the suppressor NS4B I7F in the context of pYSGR-JFH. Values represent CFU calculated from three independent transfections. Error bars represent the standard deviation from the mean. (D) Transient replication phenotypes of NS4A F48A and its suppressor NS4B I7F. Luciferase activity was measured for NS4A F48A and the suppressor NS4B I7F in the context of pYSGR-JFH/GLuc. Huh 7.5 cells were transfected with the indicated mutants, and media were collected at 6, 12, 24, 48, 72, and 96 h posttransfection. Values represent the time course of secreted GLuc activity for each transfection, expressed as an average of at least three independent transfections. Error bars represent the standard deviation from the mean.