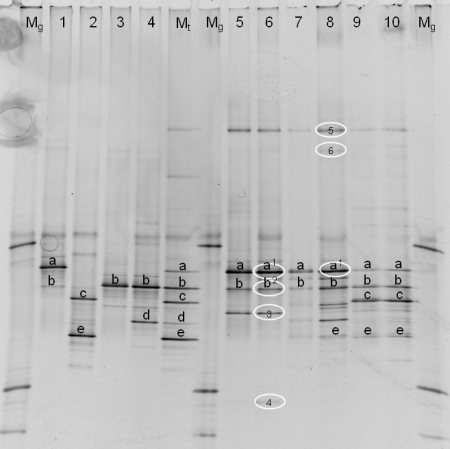
FIG. 4.
Identification of the DGGE patterns (40 to 50% denaturating gel). The patterns were obtained from milk samples taken at the end of the three simulations. Lanes: Mg, marker for analysis of the gel; 1, P. gessardii (LMG 21604T); 2, P. fragi (LMG 2191T); 3: P. lundensis (LMG 13517T); 4, P. fluorescens (LMG 1794T); Mt, type strain marker (combines lanes 1 to 4); 5, s1 (optimal storage); 6, s1 (suboptimal storage); 7, s2 (optimal storage); 8, s2 (suboptimal storage); 9, s3 (optimal storage); 10, s3 (suboptimal storage). Fragments at the same positions on the gel are marked with letters (a to e). Identifications based on sequencing of cloned fragments are encircled, as follows: 1, Pseudomonas argentinensis/Pseudomonas fulgida/Pseudomonas teesidea/P. gessardii/Pseudomonas cedrella/Pseudomonas libanensis (99.5 and 100% similarity for s1 and s2, respectively); 2, Pseudomonas vranovensis/P. lundensis (98.4% similarity); 3, Pseudomonas sp.; 4, Acinetobacter sp.; 5, P. fluorescens group (100% similarity); 6, Acinetobacter haemolyticus (99.0% similarity).