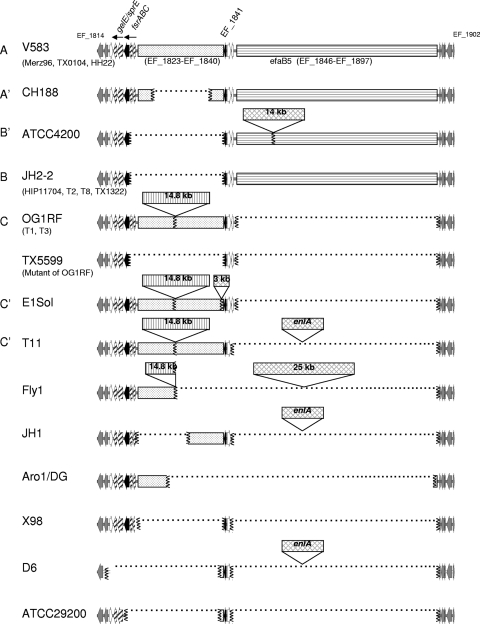
FIG. 1.
Genomic diversity of E. faecalis between EF_1814 and EF_1902. Twenty-two of the sequenced strains plus the 37-kb deletion mutant of OG1RF are shown. Redasoft Visual Cloning was used to find open reading frames (ORFs) representing EF_1814 to EF_1902 in sequences downloaded from either the NCBI server or the Broad Institute server. The first two ORFs (EF_1814 and EF_1815) and the last five ORFs (EF_1898 to EF_1902) of this region, which are common between all strains, are shown in gray. The fsrC (EF_1820) and EF_1841 genes are indicated by black arrows. When a region is missing, the junction areas are indicated with vertical squiggly lines, and the space between is filled with a dotted line. The fsrAB (EF_1821 and EF_1822), gelE (EF_1818), and sprE (EF_1817) genes are labeled with arrows filled with diagonal lines. The immediately adjacent loci, EF_1816, EF_1843, and EF_1844, are represented with white arrows. The area from EF_1823 to EF_1840 is represented with a spotted background, while the one representing the 14.8-kb region contains vertical lines. The efaB5 region from EF_1847 to EF_1896 is represented with horizontal lines. All other insertions are represented with crosshatching. Letters on the left side of the diagram indicate the genotypes based on EF_1814 to EF_1902, corresponding to Table 1.