Abstract
Lactic acid bacteria (LAB) are of industrial importance in the production of fermented foods, including sourdough-derived products. Despite their limited metabolic capacity, LAB contribute considerably to important characteristics of fermented foods, such as extended shelf-life, microbial safety, improved texture, and enhanced organoleptic properties. Triggered by the considerable amount of LAB genomic information that became available during the last decade, transcriptome and, by extension, metatranscriptome studies have become one of the most appropriate research approaches to study whole-ecosystem gene expression in more detail. In this study, microarray analyses were performed using RNA sampled during four 10-day spontaneous sourdough fermentations carried out in the laboratory with an in-house-developed LAB functional gene microarray. For data analysis, a new algorithm was developed to calculate a net expression profile for each of the represented genes, allowing use of the microarray analysis beyond the species level. In addition, metabolite target analyses were performed on the sourdough samples to relate gene expression with metabolite production. The results revealed the activation of different key metabolic pathways, the ability to use carbohydrates other than glucose (e.g., starch and maltose), and the conversion of amino acids as a contribution to redox equilibrium and flavor compound generation in LAB during sourdough fermentation.
Sourdough originates from spontaneous fermentation of a mixture of ground cereals and water (24). The sourdough microbial ecosystem is dominated by a limited number of lactic acid bacteria (LAB) and yeasts (53, 64, 72, 75, 76) and is remarkably stable over time (54), which indicates that the LAB and yeast species found in sourdough are highly adapted to this ecosystem (8, 10-12, 17, 20, 68). Adaptations that contribute to the competitive advantage of LAB in the sourdough environment are the capability to metabolize maltose, originating from the breakdown of starch by cereal amylases, by means of a maltose phosphorylase without ATP expenditure (19, 58), the use of the arginine deiminase (ADI) pathway to enhance ATP generation and improve acid tolerance (9, 15, 17, 70, 71), and the reduction of fructose to mannitol by mannitol dehydrogenase to regenerate NAD+ and to produce ATP (20, 36, 69). Despite their limited metabolic capacity, LAB contribute considerably to microbial safety, extended shelf-life, and organoleptic quality of sourdough-based food products by means of the production of organic acids, bacteriocins, exopolysaccharides (EPS), and/or flavor compounds (17, 39, 43, 55, 63). In particular, the ability to convert pyruvate and amino acids to various flavor precursors and flavor-active compounds results in an improved sensory profile for sourdough and bread (17, 25, 55). All these properties have been studied, mostly through separate pathway and gene cluster analyses, by means of physiological and molecular approaches but have never been tackled simultaneously on the ecosystem level.
LAB genome sequencing efforts over the past years (7, 31, 33, 42, 46, 47, 50, 77, 82) have resulted in the development of several whole-genome microarrays to investigate gene expression of single species (2-5, 29, 35, 37, 44, 49, 51, 52). One of these microarrays has been used to investigate the global transcriptional response of a human Lactobacillus reuteri strain to the sourdough environment, revealing that the microorganism's gene expression in this environment is changed compared to its growth in a chemically defined medium (29). However, to monitor the functional activity of LAB in complex ecosystems such as sourdough, a so-called functional gene microarray on a microbial community level was needed, allowing an overall analysis of gene expression (26, 27, 74, 83). Indeed, this kind of microarray analysis focuses on a predefined set of target genes involved in important cellular processes and metabolic reactions in the ecosystem under study. In some cases, the oligonucleotides have been designed in such a way that they are gene specific but can preferably cross-hybridize several species, thereby targeting conserved sequences shared by several species (27, 74). A LAB functional gene microarray, representing genes that play a key role in the production of carbohydrate catabolites, bacteriocins, EPS, and flavor compounds, in the stress response, and in probiotic and biosafety characteristics, as well as genes linked to negative traits, such as antibiotic resistance and virulence factors, has been developed recently (74). This microarray has been applied successfully to investigate the LAB community dynamics of two wheat and two spelt sourdough fermentations that were daily back-slopped during a period of 10 days (75).
The aim of the present study was to analyze the metatranscriptome of LAB during spontaneous sourdough fermentations, using the LAB functional gene microarray, and to relate the outcome with metabolite production by the LAB communities. In terms of proof of principle, this paper solely reports on metatranscriptomic analysis of carbohydrate utilization and amino acid metabolism.
MATERIALS AND METHODS
Sourdough fermentations.
Two wheat (D12W and D13W) and two spelt (D12S and D13S) back-slopped sourdoughs were prepared through spontaneous fermentation. Therefore, fresh 8-kg flour-water mixtures with a dough yield of 400 [(dough mass/flour mass) × 100] were incubated at 30°C for 24 h in Biostat C fermentors (Sartorius AG/B. Braun Biotech International, Melsungen, Germany). The mixture was kept homogeneous by stirring at 300 rpm. The sourdoughs were daily back-slopped over a period of 10 days by inoculating a fresh water-flour mixture with ripe sourdough (10%, wt/wt), after which the mixed dough was incubated under the same conditions as mentioned above.
RNA isolation.
RNA was isolated directly from sourdough fermentation samples to represent the metatranscriptome. Therefore, samples were taken 3 h after each refreshment step (i.e., at 27, 51, 75, 99, 123, 147, 171, 195, and 219 h), as approximately half of the acidification had taken place at this time point, indicating that the bacteria were in their exponential growth phase (64). Ten grams of sourdough was mixed with 40 ml of RNAprotect (Qiagen, Hilden, Germany) that was diluted 2:1 with phosphate-buffered saline (PBS; Invitrogen, Carslbad, CA). This mixture was kept at room temperature for 5 min, followed by centrifugation (1,000 × g for 5 min) to remove solids. The supernatant was centrifuged for a second time (5,000 × g for 15 min), and the metatranscriptome RNA was isolated from the resulting cell pellet by applying an enzymatic lysis using mutanolysin and lysozyme, after which the RNA was extracted from the resulting mixture by using an RNeasy minikit (Qiagen) following standard instructions and including mechanical disruption of the cells using glass beads, as described previously (74).
Microarray hybridization.
A LAB functional gene microarray was used that contains 2,269 oligonucleotides which target in total 406 key genes covering, among others, carbohydrate transport and metabolism, proteolysis, amino acid metabolism, and biogenic amines production and which covered as many LAB species as possible (74). The isolated metatranscriptome RNA was linearly amplified (aRNA) by using a SensAmp kit (Genisphere, Hatfield, PA), labeled with Cy3 and Cy5 dyes in a reverse transcription reaction, and hybridized for 16 h using an HS 4800 Pro automated hybridization station (Tecan Systems, Inc., San Jose, CA), as described previously (74), except that 60 pmol instead of 50 pmol of labeled aRNA was used to prepare the hybridization mixtures. The labeled aRNA of the different sourdough fermentation samples was hybridized to the microarray, using a loop design over the different time points, i.e., two consecutive samples (e.g., 27 h and 51 h, 51 h and 75 h, etc.) were hybridized on the same microarray slide, each was labeled with another fluorescent dye (Cy3 or Cy5), and the loop was closed by hybridizing the last sample (219 h) together with the first (27 h) (75).
Microarray data analysis. (i) Data preprocessing.
Given that each oligonucleotide was spotted four times on the array and each sample was hybridized twice (i.e., once in Cy3 and once in Cy5), the intensity of each oligonucleotide was measured eight times. The intensity of an oligonucleotide was considered above background level if the intensities of at least six out of eight spots were above the background level, as proposed previously (74).
To assess whether the intensity of an oligonucleotide had a significant time effect during the fermentation, analysis of variance techniques were applied for all fermentation experiments, as described previously (32, 79). Prior to the model fitting, the local background intensity was subtracted from the foreground intensity and a log2 transformation was applied. For each oligonucleotide, the average of the four Cy3 and Cy5 intensities was used. Hence, two measurements at each time point were obtained for the 2,269 oligonucleotides. In the case of a missing measurement, its value was imputed with the KNNimpute algorithm (with the number of nearest neighbors to use during imputation K set to 10 and the Euclidean distance as the distance measure), as described previously (62). A normalization model with the mixed-model approach was fit on the data set to remove dye and array effects, based on the equation: ydag = μ + Dd + (DA)da + Aa + rdag, where ydag is the log-transformed average intensity for gene g, measured on array a in dye d; μ is the global average intensity; D is the fixed dye effect with two levels (d = Cy3 or d = Cy5); A is the random array factor with nine levels (a), representing the nine arrays that were used per loop; and r is the remaining random error.
A time factor (denoted as T) with nine levels (corresponding to the time points of sampling) was fit to each gene g to assess whether the gene displayed a significant time effect, based on the following equation: rdtg = μg + Ttg + ɛdtg, where rdtg is a measure for the residuals, as obtained from the normalization equation described above for gene g at time t; μg is the intercept for gene g; T is the time effect for gene g at time t; and ɛdtg is the remaining, unexplained random error.
(ii) Calculation of net expression.
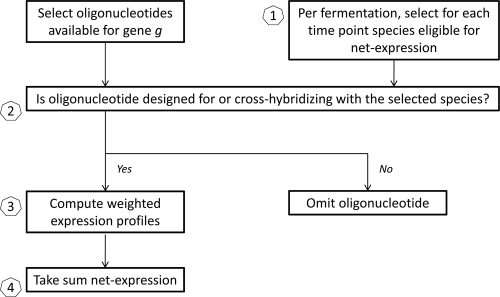
To calculate the net expression profile for each gene, thereby allowing analysis beyond the species level, an algorithm was designed, consisting of four steps (Fig. 1). First, eligible oligonucleotides were selected to calculate the net expression during the fermentations (Fig. 1, step 1). In this step, oligonucleotides that correlated to species for which none of the species-specific oligonucleotides showed hybridization intensities above background level during the fermentation process were omitted. Consequently, these species were considered to be absent in the ecosystem and thus not able to contribute to the net expression of a gene.
FIG. 1.
Schematic overview of the net expression algorithm.
This decision was refined by taking false-positive hybridization intensities into account, which were obtained during validation hybridizations using RNA from 18 LAB strains (74). Briefly, for each of the 46 LAB species represented on the microarray, oligonucleotides related to the considered species that showed hybridization intensities above background level when RNA from other species was hybridized were selected. Subsequently, the hybridization intensities of these selected (false-positive) oligonucleotides were used to calculate a one-sided 99% confidence interval for that considered species. For all time points in the current study, a species was considered present in the ecosystem at that time point, and thus able to contribute to the net expression, in cases where the number of oligonucleotides with hybridization intensities above background level was greater than the upper limit of the calculated confidence interval. As a result, the remaining oligonucleotides that showed no hybridization intensity above background level could be correctly interpreted as being correlated to a nonexpressed gene of a species that was present in the ecosystem or as an oligonucleotide that was not well designed.
In the second step (Fig. 1), poor performing species-specific oligonucleotides were identified. Indeed, data obtained during the validation of the microarray using DNA from 18 LAB strains were taken into account, i.e., information on 86% of the 2,269 oligonucleotides (74). Oligonucleotides that showed a hybridization intensity that was not above background level were considered technically not suitable and were omitted. After the first two steps, an oligonucleotide passing both steps and having a hybridization intensity that was not above background level could be interpreted as true negative, i.e., representing a gene that was not expressed by the corresponding species at the given time point in the ecosystem.
In the third step (Fig. 1), weights were assigned to the oligonucleotides that passed the previous two steps, thereby taking into account possible cross-hybridization. In cases in which a gene was represented by only one oligonucleotide, designed for or likely to cross-hybridize with a given species present in the ecosystem at a specific fermentation stage, the oligonucleotide obtained a weight equal to 1. In the case of a gene that was represented by more than one oligonucleotide for a given species, each oligonucleotide was assigned a weight equal to 1/n, where n was the number of oligonucleotides for the gene in that specific species.
In the fourth step (Fig. 1), the net expression was calculated for each gene at each time point during fermentation by taking the log of the sum of the weighted intensities for each species considered present in the ecosystem. For time points where all oligonucleotides were omitted in the first two steps, information was insufficient and net expression could not be calculated. The net expression profile, being a measure for gene expression, as well as the intensity profiles of the contributing oligonucleotides were plotted as a function of time. Expression was considered physiologically meaningful if the expression level was above a threshold of 1, corresponding to an intensity that was 10 times higher than the background level.
Metabolite target analysis.
Maltose concentrations (in g kg−1) were determined by high-performance anion-exchange chromatography with pulsed amperometric detection on an ICS-3000 chromatograph (Dionex, Sunnyvale, CA), as described previously (64). Concentrations of the amino acid metabolites 2-methyl-propanol, 2-methyl-butanol, and 3-methyl-butanol, expressed in arbitrary units, as 100 × [(peak area for the compound)/(peak area of the internal standard × g of sample)], were determined by static headspace gas chromatography-mass spectrometry analysis on an Agilent 6890 gas chromatograph coupled to an Agilent 5973N mass spectrometer (Agilent Technologies, Palo Alto, CA), as described previously (64).
Microarray data accession numbers.
The microarray data were deposited in the NCBI Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/) under accession numbers GPL5459 (microarray including detailed annotation), GSE15692 (D12S), GSE15686 (D12W), GSE15691 (D13S), and GSE15693 (D13W).
RESULTS
The metatranscriptome RNA samples were hybridized to the LAB functional gene microarray to reveal the expression patterns of genes involved in carbohydrate utilization and amino acid metabolism during 10-day back-slopped sourdough fermentations. No samples were available for day 2 (51 h) and day 3 (75 h) of fermentation D13S due to poor-quality RNA. Although this LAB functional gene microarray was intended to be used beyond the species level, only 15.5% of the 2,269 oligonucleotides were species independent, while the remaining oligonucleotides could hybridize only one species. This was mainly due to the limited availability of gene sequences at the time of design and production (74). As a result, gene expression still could be linked to specific LAB species in the sourdough fermentation process (75), which allowed us to include species information in this study for ease of understanding.
Phosphotransferase transport system.
On the microarray, four genes involved in carbohydrate transport were represented. None of the sourdough fermentations showed expression of the lactose phosphotransferase transport system (PTS). The gene coding for a maltose/maltodextrine ABC transporter, which was represented by oligonucleotides designed based on sequences from Lactobacillus plantarum and Lactococcus lactis showed a low net expression in fermentation D13W, originating from the expression by Lc. lactis during the first 2 days of back-slopping. For the other fermentations, the net expression level was below the expression level threshold. Yet, carbohydrate transport occurred. A PEP-carbohydrate phosphotransferase system enzyme I showed a high net expression in all sourdough fermentations and was represented by oligonucleotides related to Lactobacillus sakei, Lactobacillus brevis, L. plantarum, and Lc. lactis. Also, a phosphocarrier protein showed high net expression in all fermentation steps during the whole 10-day back-slopping period.
Carbohydrate metabolism.
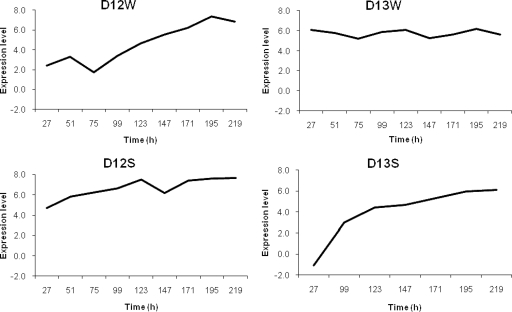
Ninety-five genes involved in carbohydrate metabolism were represented on the microarray. All genes encoding enzymes involved in the conversion of glucose into pyruvate through glycolysis, such as fructose 1,6-bisphosphate aldolase, triose phosphate isomerase, glyceraldehyde 3-phosphate dehydrogenase, phosphoglycerokinase, and pyruvate kinase, showed a high net expression in all fermentations during the whole sourdough back-slopping period. Also, the genes encoding glucose 6-phosphate dehydrogenase and 6-phosphogluconate dehydrogenase of the phosphoketolase pathway showed a high net expression in all fermentations, encompassing the gene expression levels in L. plantarum, Pediococcus pentosaceus, and Lactobacillus curvatus. The expression level of the oligonucleotide related to L. curvatus was the highest, perhaps caused by cross-hybridization with L. plantarum, as indicated by the oligonucleotide annotation. Additionally, transketolase gene expression was only found in Lc. lactis in the wheat fermentations. None of the sourdough fermentations showed transaldolase expression. Genes encoding enzymes involved in the pentose phosphate pathway, such as ribulose 5-phosphate 3-epimerase and ribose 5-phosphate isomerase, were moderately to highly expressed by P. pentosaceus and L. plantarum, respectively, in all fermentations. Further, citrate permease was not expressed at all, citrate lyase was only expressed by L. plantarum, succinate dehydrogenase was only expressed by P. pentosaceus, fumarase was expressed moderately and was linked with P. pentosaceus and L. plantarum, and pyruvate formate lyase was highly expressed by Lc. lactis only in the wheat fermentations and by L. sakei only in the spelt fermentations. The moderate (D12W) to high (D13W) net expression of the catabolite control protein (CcpA) gene was linked with Lc. lactis during the first days of back-slopping during the wheat fermentations and with L. plantarum in all fermentations (Fig. 2).
FIG. 2.
Net expression profiles for the catabolite control protein gene in fermentations D12W, D13W, D12S, and D13S.
The gene encoding fructokinase, which is involved in fructose degradation, showed a high net expression in all sourdough fermentations. This expression was linked with Lactobacillus fermentum in fermentations D12W and D12S and with L. plantarum in fermentations D13W and D13S, although the contribution of the latter LAB species was only moderate. The gene encoding β-fructofuranosidase, which is involved in sucrose and oligofructose degradation, showed a high net expression in all sourdough fermentations linked with L. plantarum. β-Galactosidase, which is involved in the degradation of β-galactosides such as lactose, showed a moderate net expression that was linked with L. plantarum and P. pentosaceus. α-Amylase, involved in starch degradation, was moderately expressed by L. plantarum only in the two wheat fermentations; the expression of this gene in the two spelt fermentations was hardly above the expression level threshold.
Maltose phosphorylase.
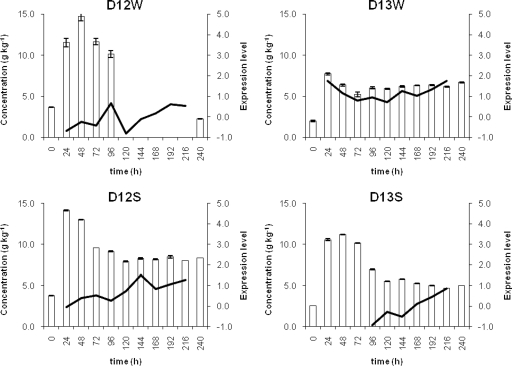
Ten oligonucleotides represented the enzyme maltose phosphorylase on the microarray. For fermentations D12W and D13S, the net expression of the coding gene was below the expression level threshold (Fig. 3). For fermentation D12S, its net expression was slightly above the threshold during the last 4 days of back-slopping and was linked with L. plantarum. For fermentation D13W, its net expression was low but above the threshold for almost the whole 10-day back-slopping period and was linked with L. plantarum and Lc. lactis. The largest amounts of maltose were reached on days 2 and 3 of back-slopping in all sourdough fermentations (Fig. 3). Except for fermentation D13W, which showed little variation in maltose concentration over the 10-day back-slopping period, the concentration decreased after day 3.
FIG. 3.
Relation between the net expression of the maltose phosphorylase gene (black line; right axes) and the concentration of maltose (white bars; left axes) in fermentations D12W, D13W, D12S, and D13S. No data on the maltose concentration were available for time points 120 h, 144 h, 168 h, 192 h, or 216 h for fermentation D12W.
Proteolysis and amino acid conversions.
One hundred genes involved in proteolysis and amino acid conversions were represented on the microarray. Net expression profiles of genes encoding peptide transport systems, such as the genes coding for the subunits of the oligopeptide transport system (oppBCDF) and the gene encoding a di/tripeptide transporter (dtpT), showed moderate expression, originating from L. plantarum in all sourdough fermentations, from Lc. lactis in the two wheat fermentations, and from L. sakei in the two spelt fermentations. The subunits for the oligopeptide ABC transporter (optABCDF) were only expressed by Lc. lactis in the two wheat fermentations.
Net expression profiles of genes encoding intracellular degradation of peptides, such as those coding for nonspecific aminopeptidases, dipeptidases, tripeptidases, X-prolyl-dipeptidyl aminopeptidases, and several endopeptidases, showed expression by L. plantarum in all sourdough fermentations and by Lc. lactis in the two wheat fermentations. No expression was found for the cell wall-associated serine proteinase, the only proteinase represented on the microarray.
Net expression profiles of the gene coding for the enzyme aromatic aminotransferase, which catalyzes the transamination step in the conversion of the aromatic amino acids tyrosine, phenylalanine, and tryptophan to their corresponding α-keto acids, showed moderate expression by L. plantarum in all sourdough fermentations and by Lc. lactis in the two wheat fermentations. Genes encoding enzymes in the aromatic amino acid biosynthesis pathway were moderately expressed by L. plantarum in some sourdough fermentations, such as the genes coding for 3-dehydroquinate dehydratase, shikimate 5-dehydrogenase, shikimate kinase, and chorismate synthase.
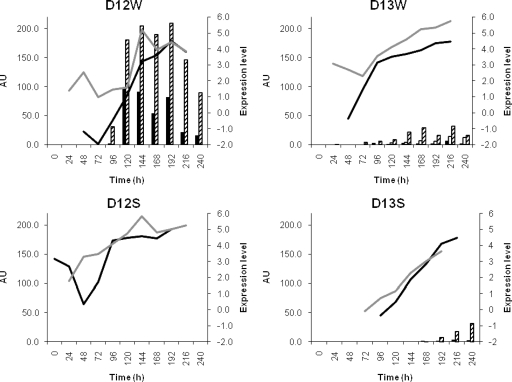
Several genes encoding enzymes involved in different glutamate-related pathways were expressed during the sourdough fermentations. Expression of a glutamate dehydrogenase gene was found solely for L. plantarum in all fermentations. The gene coding for the branched-chain aminotransferase, which catalyzes the first step in the conversion of the branched-chain amino acids valine, leucine, and iso-leucine into their corresponding α-keto acids, was expressed by L. plantarum in all fermentations and additionally by Lc. lactis in fermentation D13W (Fig. 4). A simultaneous increase in the production of several amino acid metabolites was observed together with the expression of the glutamate dehydrogenase gene and the branched-chain aminotransferase gene, which was reflected in the concentrations of 2-methyl-propanol, 2-methyl-butanol, and 3-methyl-butanol (Fig. 4). The gene coding for aspartate aminotransferase, involved in the metabolism of aspartate, was expressed by L. plantarum in all fermentations and by P. pentosaceus in fermentations D12W, D12S, and D13S.
FIG. 4.
Relation between the net expression of the glutamate dehydrogenase gene (black line; right axes) and the branched-chain aminotransferase gene (gray line; right axes) and the production of the branched-chain amino acid metabolites 2-methyl-propanol (black bars; left axes), 2-methyl-butanol (white bars; left axes), and 3-methyl-butanol (shaded bars; left axes) in fermentations D12W, D13W, D12S, and D13S. No data on the concentrations of these alcohols were available for time points 120 h, 144 h, 168 h, 192 h, or 216 h for fermentation D12W. AU, arbitrary units, determined as 100 × [(peak area for compound)/(peak area for internal standard × g of sample)].
Arginine deiminase pathway.
All four genes involved in the ADI pathway were represented on the microarray. The gene coding for the arginine/ornithine antiporter was only expressed by Lc. lactis in fermentation D13W during days 2 and 3 of the back-slopping process. The gene expression at other days and in other sourdough fermentations was below the expression level threshold. The net expression of the gene coding for arginine deiminase was linked with high expression by Lc. lactis during the first days of back-slopping during fermentations D12W and D13W and from Enterococcus faecium in the first days of back-slopping during fermentation D12S. Its net expression in fermentation D13S did not reach the threshold. The gene coding for ornithine carbamoyltransferase showed a moderate net expression over the whole back-slopping period in fermentations D12W, D13W, and D12S. In fermentation D13S, the net expression was above the expression level threshold during the last 3 days. The gene coding for carbamate kinase showed moderate to high expression by Lc. lactis only during the first days of back-slopping in fermentations D12W and D13W, although an additional weak expression by P. pentosaceus in fermentation D12W was noticed. Further, weak expression of this gene by E. faecium during the first day of back-slopping during fermentation D12S was observed. In fermentation D13S, no expression of this gene could be detected.
Decarboxylation of amino acids.
The gene gadB, coding for the enzyme glutamate decarboxylase, was moderately expressed by L. plantarum only in fermentations D12W and D13S during the last days of back-slopping. A higher net expression was observed in fermentation D12S from day 3 on and also by L. plantarum, as well as in fermentation D13W, where the gene was moderately expressed by Lc. lactis during the first 3 days of back-slopping and by L. plantarum from day 5 on. The gene tyrDC, coding for the enzyme tyrosine decarboxylase, showed high net expression during the first 2 to 3 days of back-slopping in fermentations D12W, D13W, and D12S. The oligonucleotide related to this gene was designed based on a sequence from Enterococcus faecalis and could, according to information gathered during oligonucleotide design, cross-hybridize with Lc. lactis. No expression was observed in fermentation D13S. No other decarboxylase gene was expressed in any of the sourdough fermentations.
DISCUSSION
Functional gene expression is usually studied by applying real-time PCR for specific genes and/or (meta)transcriptome analysis with whole-genome microarrays representing all protein-coding genes of a particular genome (3, 29). Real-time PCR is, however, limited to investigations in which the sequences of the genes of interest are known, since mismatching primers result in nonefficient PCR assays and hence hamper proper data interpretation. Consequently, real-time PCR is not the best-suited approach in metatranscriptome analyses. Until now, the use of functional gene microarrays has been restricted to the analysis of environmental samples, such as soil (27), feces (34), and sludge (26). Fermented food ecosystems are in general of moderate complexity, compared to soil or the human colon, and their microbial species diversity and community dynamics are often well understood (16, 30, 80), allowing functional gene microarray analysis of these food ecosystems beyond the species level.
In this study, a LAB functional gene microarray was used to monitor global gene expression of sourdough fermentation processes with daily back-slopping. The microarray was designed to assess global gene expression of 406 target genes in a species-independent way, thereby focusing on gene function rather than on the species present (74). As this is a new focus for the use of microarrays and the analysis of hybridization data, a successful method was set up to calculate a measure for the net expression of genes, i.e., the global level of expression per gene function during fermentation, regardless of the species present. These net expression profiles were used to further understand spontaneous wheat and spelt sourdough fermentations.
A highly efficient use of the available nutrients is a prerequisite for LAB to dominate a certain ecosystem. Wheat and spelt flours are rich in carbohydrates, peptides and amino acids, minerals, vitamins, and other growth factors. Carbohydrates are the main energy source for LAB, and both the glycolytic pathway and the phosphoketolase pathway were active during the sourdough fermentations performed, indicating the breakdown of both hexoses and pentoses in these LAB species (8). Further, it has been shown that the phosphoketolase bypass is activated in L. plantarum in response to lactic acid stress (51). Wheat and spelt flours contain starch-derived maltose, sucrose, and fructose as freely available carbohydrates. Starch is the main carbohydrate source in wheat flour and is degraded by endogenous amylases. The ability to hydrolyze starch, either by cereal enzymes or by enzymes secreted by microorganisms, is known to be an important prerequisite in sourdough ecosystems. The α-amylase-encoding gene was only moderately expressed by Lc. lactis in the beginning of the two wheat fermentations, although amylolytic strains of Lc. lactis, L. plantarum, and L. fermentum have been found before in cereal fermentations (13, 18). Furthermore, the sourdough microbiota apparently exhibited the capacity to degrade other carbohydrates relevant for sourdough, such as sucrose and fructose, indicating that a high degree of flexibility toward the available carbohydrates is necessary for successful competition (76). If excessive glucose is present, expression of genes encoding enzymes involved in the metabolism of other carbohydrates is often repressed through carbon catabolite repression, which is mediated via the catabolite control protein CcpA (41, 45, 48, 61). The apparent high expression of the gene coding for the CcpA protein by L. plantarum in all four sourdough fermentations studied could indicate repression of genes coding for such enzymes involved in the metabolism of carbohydrates other than glucose.
Although typical members of sourdough environments are characterized by their ability to use maltose as carbohydrate source, the gene coding for maltose phosphorylase was only moderately expressed by L. plantarum, indicating that the usage of maltose as carbohydrate source by L. plantarum was not preferential, which corresponded with the high expression of the gene coding for CcpA. As no L. fermentum-related oligonucleotides for the maltose phosphorylase gene were available on the microarray, due to a lack of sequence information at the time of oligonucleotide design and microarray production, gene expression linked to L. fermentum cannot be reported. However, because L. fermentum is known to be an important member of the sourdough ecosystem and is able to use maltose as energy source (64), it is likely that L. fermentum plays an important role in maltose consumption (69).
Differences between wheat and spelt sourdoughs in gene expression levels involved in carbohydrate metabolism could be mainly ascribed to gene expression linked with Lc. lactis, which was almost exclusively observed in the wheat sourdoughs. This is exemplified by the expression of a transketolase gene by Lc. lactis. This enzyme could be involved in a less common pathway for xylose fermentation in Lc. lactis, yielding relatively more lactate (59) and indicating heterofermentation by this generally accepted homofermentative LAB species (6), or in a Bifidobacterium-like phosphoketolase bypass leading to acetate production, as has been demonstrated in L. plantarum (51). This bypass also requires the action of transaldolase, which did not show expression levels above background, possibly indicating that only pentoses were routed through the phosphoketolase pathway in L. plantarum. Additionally, a moderate β-galactosidase gene expression level (in L. plantarum, Lc. lactis, and P. pentosaceus), as observed in the present study, implies possible degradation of the raffinose or stachyose present in wheat flour (56). The expression of genes involved in xylose utilization indicates breakdown of components from plant heteropolysaccharides, such as arabinoxylans, that undergo degradation by cereal enzymes (22). The use of these carbohydrates may therefore give a competitive advantage to the corresponding bacteria, explaining why they can become dominant during a back-slopped sourdough fermentation process.
Efficient nitrogen metabolism is another prerequisite for dominating sourdough LAB species. In general, peptides present in a food matrix (e.g., milk) are taken up by LAB and are degraded intracellularly (38). The resulting amino acids may be used for biosynthesis or other metabolic activities (12, 17, 65). In the case of sourdough LAB, in particular, it has been shown that accumulated amino acids that are not further metabolized, such as phenylalanine, are transported out of the cell (66). In the present study, no expression of the only proteinase represented on the microarray was found, namely, of the cell wall-associated serine proteinase gene. This confirmed that proteinase activity might not be of utmost importance for the microbiota present, as a sufficient amount of peptides is already generated by the action of flour proteinases (65, 76). However, the LAB functional gene microarray did not contain oligonucleotides targeting this gene in L. plantarum or L. fermentum, and so no clear conclusion can be drawn regarding the importance of extracellular proteolytic activity for competitiveness, although strains of these species elaborating proteinase activities have been isolated from sourdough (14, 21). The expression of many peptidase-encoding genes, observed during the sourdough fermentations carried out, underline the importance of adaptiveness and flexibility for successful competition of LAB in a sourdough ecosystem.
The importance of the ADI pathway with respect to acid stress and energy generation (9, 15, 40) was not clear. Indeed, the LAB functional gene microarray analysis revealed that only some genes of this pathway were expressed during the first days of back-slopping by different species and their expression decreased later on. This might be peculiar, because stress resistance is of high importance in order to dominate a fermented food ecosystem. However, it has been shown that a sourdough-related strain of L. fermentum efficiently converts arginine through the ADI pathway from the late exponential phase on (70, 71). Unfortunately, no L. fermentum-related oligonucleotides for the ADI pathway genes were available on the microarray, which may explain the lack of ADI expression. Also, the sampling point, i.e., 3 h after back-slopping, might have been too early to observe gene expression for the genes involved in the ADI pathway, as the conversion of arginine into citrulline and ornithine typically starts in the second half of the exponential growth phase. Therefore, it has been suggested that a critical cell density, dependent on the environmental pH, must be reached before the start of arginine consumption. Indeed, although the ADI genes are not regulated by environmental pH, it has been observed that the consumption of arginine starts at a lower cell density, thus earlier in the exponential growth phase, when the environmental pH is lower (70).
Amino acid conversions often use NADH plus H+ or NAD+ as cofactors for their redox reactions, whatever is needed, contributing to competitiveness (1). This was exemplified by the expression of the genes coding for enzymes of the shikimate pathway. Although this pathway is referred to as the aromatic amino acid biosynthetic pathway, it might also play a role in redox balancing (28, 78), allowing the production of hydroxyl acids or alcohols. In addition, many flavor compounds originate from amino acid conversions. They are initiated by the generation of the corresponding α-keto acids through transamination with α-ketoglutarate as the most important amino acceptor. The amount of α-ketoglutarate is known to be the limiting factor in flavor formation (57, 81). There are three known ways for generating α-ketoglutarate: (i) from glutamate through the action of an NAD+-dependent glutamate dehydrogenase; (ii) from citrate through the action of citrate lyase and aspartate aminotransferase; and (iii) from either pyruvate or citrate through the oxidative part of the citric acid cycle. This last possibility is often not active in LAB (60). In the present study, a gene coding for glutamate dehydrogenase was highly expressed. However, as the glutamate dehydrogenase enzymes can be divided into two classes, i.e., NAD+-dependent and NADP+-dependent glutamate dehydrogenases (23), it remains unclear to which class the expressed gene belongs, especially because the behavior is strain dependent. Further, no decisive answer could be given concerning the involvement of the oxidative branch of the citric acid cycle, relying on the enzymes pyruvate carboxylase, citrate synthase, aconitase, and isocitrate dehydrogenase (73). Of these enzymes, only the pyruvate carboxylase gene was represented on the microarray, and it was highly expressed. Also, the genes coding for citrate lyase and aspartate aminotransferase were highly expressed, but no expression was observed for the citrate permease gene, and so the route starting from citrate to replenishing the pool of α-ketoglutarate remains questionable. However, as it has been shown that glutamate, which is the most abundant amino acid in wheat proteins, can be obtained by intracellular conversion of glutamine by glutaminase by cereal-associated LAB, glutamate could still be the source of α-ketoglutarate in this sourdough environment, indicating a route to an improved flavor profile (67). Furthermore, the fact that the expression of pyruvate-formate lyase was only above background during the first days of back-slopping, as a result of expression by Lc. lactis and L. sakei, indicates that homolactic fermentation was the main energy-generating pathway in L. plantarum and that mixed-acid fermentation was not elicited under the nonetheless-stressful conditions of the sourdough ecosystem.
To conclude, the LAB functional gene microarray was successfully used to follow global gene expression of two wheat and two spelt sourdough fermentations that were daily sampled during a 10-day sourdough fermentation process with daily back-slopping. The LAB species present in the sourdoughs employed many different mechanisms for efficient use of the available nutrients, such as the use of carbohydrates other than glucose (e.g., starch and maltose) and the conversion of amino acids (contributing to the redox equilibrium and generating flavor compounds). Also, the data indicated that this LAB functional gene microarray is a dynamic tool, and it will need regular updates of the cross-hybridization annotation of the current oligonucleotides as well as the design of additional oligonucleotides with respect to new LAB genome information as it becomes available in the public domain.
Acknowledgments
This work was financed by SBO project IWT-030263 of the Institute for the Promotion of Innovation by Science and Technology in Flanders, Belgium (IWT-Vlaanderen). S.W. and L.D.V. acknowledge their financial support from the Research Council of the Vrije Universiteit Brussel (BOF, GOA, and IOF projects). G.H. is a postdoctoral fellow of the Fund for Scientific Research Flanders, Belgium (FWO-Vlaanderen).
We are grateful to Kizi Coeck, Ruth Maes, Ilse Scheirlinck, and Ann Van Schoor for their contributions.
Footnotes
Published ahead of print on 19 November 2010.
REFERENCES
- 1.Ardö, Y. 2006. Flavour formation by amino acid catabolism. Biotech. Adv. 24:238-242. [DOI] [PubMed] [Google Scholar]
- 2.Azcarate-Peril, M. A., et al. 2005. Microarray analysis of a two-component regulatory system involved in acid resistance and proteolytic activity in Lactobacillus acidophilus. Appl. Environ. Microbiol. 71:5794-5804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Azcarate-Peril, M. A., R. Tallon, and T. R. Klaenhammer. 2009. Temporal gene expression and probiotic attributes of Lactobacillus acidophilus during growth in milk. J. Dairy Sci. 92:870-886. [DOI] [PubMed] [Google Scholar]
- 4.Barrangou, R., et al. 2006. Global analysis of carbohydrate utilization by Lactobacillus acidophilus using cDNA microarrays. Proc. Natl. Acad. Sci. U. S. A. 103:3816-3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berger, B., et al. 2007. Similarity and differences in the Lactobacillus acidophilus group identified by polyphasic analysis and comparative genomics. J. Bacteriol. 189:1311-1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bolotin, A., et al. 2001. The complete genome sequence of the lactic acid bacterium Lactococcus lactis ssp. lactis IL1403. Genome Res. 11:731-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Callanan, M., et al. 2008. Genome sequence of Lactobacillus helveticus, an organism distinguished by selective gene loss and insertion sequence element expansion. J. Bacteriol. 190:727-735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Corsetti, A., and L. Settanni. 2007. Lactobacilli in sourdough fermentation. Food Res. Int. 40:539-558. [Google Scholar]
- 9.De Angelis, M., et al. 2002. Arginine catabolism by sourdough lactic acid bacteria: purification and characterization of the arginine deiminase pathway enzymes from Lactobacillus sanfranciscensis CB1. Appl. Environ. Microbiol. 68:6193-6201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Vuyst, L., and P. Neysens. 2005. The sourdough microflora: biodiversity and metabolic interactions. Trends Food Sci. Technol. 16:43-56. [Google Scholar]
- 11.De Vuyst, L., and M. Vancanneyt. 2007. Biodiversity and identification of sourdough lactic acid bacteria. Food Microbiol. 24:120-127. [DOI] [PubMed] [Google Scholar]
- 12.De Vuyst, L., G. Vrancken, F. Ravyts, T. Rimaux, and S. Weckx. 2009. Biodiversity, ecological determinants, and metabolic exploitation of sourdough microbiota. Food Microbiol. 26:666-675. [DOI] [PubMed] [Google Scholar]
- 13.Diaz-Ruiz, G., J. P. Guyot, F. Ruiz-Teran, J. Morlon-Guyot, and C. Wacher. 2003. Microbial and physiological characterization of weakly amylolytic but fast-growing lactic acid bacteria: a functional role in supporting microbial diversity in pozol, a Mexican fermented maize beverage. Appl. Environ. Microbiol. 69:4367-4374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Di Cagno, R., et al. 2002. Proteolysis by sourdough lactic acid bacteria: effects on wheat flour protein fractions and gliadin peptides involved in human cereal intolerance. Appl. Environ. Microbiol. 68:623-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fernández, M., and M. Zúñiga. 2006. Amino acid catabolic pathways of lactic acid bacteria. Crit. Rev. Microbiol. 32:155-183. [DOI] [PubMed] [Google Scholar]
- 16.Fleet, G. H. 1999. Microorganisms in food ecosystems. Int. J. Food Microbiol. 50:101-117. [DOI] [PubMed] [Google Scholar]
- 17.Gänzle, M. G., N. Vermeulen, and R. F. Vogel. 2007. Carbohydrate, peptide and lipid metabolism of lactic acid bacteria in sourdough. Food Microbiol. 24:128-138. [DOI] [PubMed] [Google Scholar]
- 18.Giraud, E., A. Champailler, and M. Raimbault. 1994. Degradation of raw starch by a wild amylolytic strain of Lactobacillus plantarum. Appl. Environ. Microbiol. 60:4319-4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gobbetti, M. 1998. Interactions between lactic acid bacteria and yeasts in sourdoughs. Trends Food Sci. Technol. 9:267-274. [Google Scholar]
- 20.Gobbetti, M., M. De Angelis, A. Corsetti, and R. Di Cagno. 2005. Biochemistry and physiology of sourdough lactic acid bacteria. Trends Food Sci. Technol. 16:57-69. [Google Scholar]
- 21.Gobbetti, M., E. Smacchi, and A. Corsetti. 1996. The proteolytic system of Lactobacillus sanfrancisco CB1: purification and characterization of a proteinase, a dipeptidase, and an aminopeptidase. Appl. Environ. Microbiol. 62:3220-3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goesaert, H., et al. 2005. Wheat flour constituents: how they impact bread quality, and how to impact their functionality. Trends Food Sci. Technol. 16:12-30. [Google Scholar]
- 23.Gutiérrez-Méndez, N., E. Valenzuela-Soto, A. F. González-Córdova, and B. Vallejo-Cordoba. 2008. α-Ketoglutarate biosynthesis in wild and industrial strains of Lactococcus lactis. Lett. Appl. Microbiol. 47:202-207. [DOI] [PubMed] [Google Scholar]
- 24.Hammes, W. P., and M. G. Gänzle. 1998. Sourdough breads and related products, p. 199-216. In B. J. B. Wood (ed.) Microbiology of fermented foods. Blackie Academic and Professional, London, United Kingdom.
- 25.Hansen, A., and P. Schieberle. 2005. Generation of aroma compounds during sourdough fermentation: applied and fundamental aspects. Trends Food Sci. Technol. 16:85-94. [Google Scholar]
- 26.He, S., et al. 2010. Metatranscriptomic array analysis of ‘Candidatus Accumulibacter phosphatis’-enriched enhanced biological phosphorus removal sludge. Environ. Microbiol. 12:1205-1217. [DOI] [PubMed] [Google Scholar]
- 27.He, Z., et al. 2007. GeoChip: a comprehensive microarray for investigating biogeochemical, ecological and environmental processes. ISME J. 1:67-77. [DOI] [PubMed] [Google Scholar]
- 28.Herrmann, K. M. 1995. The shikimate pathway as an entry to aromatic secondary metabolism. Plant Physiol. 107:7-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hüfner, E., R. A. Britton, S. Roos, H. Jonsson, and C. Hertel. 2008. Global transcriptional response of Lactobacillus reuteri to the sourdough environment. Syst. Appl. Microbiol. 31:323-338. [DOI] [PubMed] [Google Scholar]
- 30.Hutkins, R. W. 2006. Microbiology and technology of fermented foods. Blackwell Publishing, Ames, IA.
- 31.Kankainen, M., et al. 2009. Comparative genomic analysis of Lactobacillus rhamnosus GG reveals pili containing a human mucus binding protein. Proc. Natl. Acad. Sci. U. S. A. 106:17193-17198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kerr, M. K., M. Martin, and G. A. Churchill. 2000. Analysis of variance for gene expression microarray data. J. Comput. Biol. 7:819-837. [DOI] [PubMed] [Google Scholar]
- 33.Kim, J. F., et al. 2008. The complete genome sequence of Leuconostoc citreum KM20. J. Bacteriol. 190:3093-3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klaassens, E. S., et al. 2009. Mixed-species genomic microarray analysis of fecal samples reveals differential transcriptional responses of bifidobacteria in breast- and formula-fed infants. Appl. Environ. Microbiol. 75:2668-2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kok, J., G. Buist, A. L. Zorner, S. van Hijum, and O. P. Kuipers. 2005. Comparative and functional genomics of lactococci. FEMS Microbiol. Rev. 29:411-433. [DOI] [PubMed] [Google Scholar]
- 36.Korakli, M., and R. F. Vogel. 2003. Purification and characterisation of mannitol dehydrogenase from Lactobacillus sanfranciscensis. FEMS Microbiol. Lett. 220:281-286. [DOI] [PubMed] [Google Scholar]
- 37.Kuipers, O. P., et al. 2002. Transcriptome analysis and related databases of Lactococcus lactis. Antonie Van Leeuwenhoek 82:113-122. [PubMed] [Google Scholar]
- 38.Kunji, E. R. S., I. Mierau, A. Hagting, B. Poolman, and W. N. Konings. 1996. The proteolytic systems of lactic acid bacteria. Antonie Van Leeuwenhoek 70:187-221. [DOI] [PubMed] [Google Scholar]
- 39.Leroy, F., and L. De Vuyst. 2004. Lactic acid bacteria as functional starter cultures for the food fermentation industry. Trends Food Sci. Technol. 15:67-78. [Google Scholar]
- 40.Lu, C.-D. 2006. Pathways and regulation of bacterial arginine metabolism and perspectives for obtaining arginine overproducing strains. Appl. Microbiol. Biotechnol. 70:261-272. [DOI] [PubMed] [Google Scholar]
- 41.Luesink, E. J., O. P. Kuipers, and W. M. de Vos. 1998. Regulation of the carbohydrate metabolism in Lactococcus lactis and other lactic acid bacteria. Lait 78:69-76. [Google Scholar]
- 42.Mazé, A., et al. 2010. Complete genome sequence of the probiotic Lactobacillus casei strain BL23. J. Bacteriol. 192:2647-2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Messens, W., and L. De Vuyst. 2002. Inhibitory substances produced by Lactobacilli isolated from sourdoughs: a review. Int. J. Food Microbiol. 72:31-43. [DOI] [PubMed] [Google Scholar]
- 44.Molenaar, D., et al. 2005. Exploring Lactobacillus plantarum genome diversity by using microarrays. J. Bacteriol. 187:6119-6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Monedero, V., M. J. Gosalbes, and G. Pérez-Martínez. 1997. Catabolite repression in Lactobacillus casei ATCC 393 is mediated by CcpA. J. Bacteriol. 179:6657-6664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Morita, H., et al. 2008. Comparative genome analysis of Lactobacillus reuteri and Lactobacillus fermentum reveal a genomic island for reuterin and cobalamin production. DNA Res. 15:151-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Morita, H., et al. 2009. Complete genome sequence of the probiotic Lactobacillus rhamnosus ATCC 53103. J. Bacteriol. 191:7630-7631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Muscariello, L., R. Marasco, M. De Felice, and M. Sacco. 2001. The functional ccpA gene is required for carbon catabolite repression in Lactobacillus plantarum. Appl. Environ. Microbiol. 67:2903-2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pavlovic, M., S. Hormann, R. F. Vogel, and M. A. Ehrmann. 2005. Transcriptional response reveals translation machinery as target for high pressure in Lactobacillus sanfranciscensis. Arch. Microbiol. 184:11-17. [DOI] [PubMed] [Google Scholar]
- 50.Pfeiler, E. A., and T. R. Klaenhammer. 2007. The genomics of lactic acid bacteria. Trends Microbiol. 15:546-553. [DOI] [PubMed] [Google Scholar]
- 51.Pieterse, B., R. J. Leer, F. H. J. Schuren, and M. J. van der Werf. 2005. Unravelling the multiple effects of lactic acid stress on Lactobacillus plantarum by transcription profiling. Microbiology 151:3881-3894. [DOI] [PubMed] [Google Scholar]
- 52.Santos, F., et al. 2009. Effect of amino acid availability on vitamin B-12 production in Lactobacillus reuteri. Appl. Environ. Microbiol. 75:3930-3936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Scheirlinck, I., et al. 2007. Influence of geographical origin and flour type on diversity of lactic acid bacteria in traditional Belgian sourdoughs. Appl. Environ. Microbiol. 73:6262-6269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Scheirlinck, I., et al. 2008. Taxonomic structure and stability of the bacterial community in Belgian sourdough ecosystems as assessed by culture and population fingerprinting. Appl. Environ. Microbiol. 74:2414-2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schnürer, J., and J. Magnusson. 2005. Antifungal lactic acid bacteria as biopreservatives. Trends Food Sci. Technol. 16:70-78. [Google Scholar]
- 56.Silvestroni, A., C. Connes, F. Sesma, G. S. de Giori, and J.-C. Piard. 2002. Characterization of the melA locus for α-galactosidase in Lactobacillus plantarum. Appl. Environ. Microbiol. 68:5464-5471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Smit, G., B. A. Smit, and W. J. M. Engels. 2005. Flavour formation by lactic acid bacteria and biochemical flavor profiling of cheese products. FEMS Microbiol. Rev. 29:591-610. [DOI] [PubMed] [Google Scholar]
- 58.Stolz, P., W. P. Hammes, and R. F. Vogel. 1996. Maltose-phosphorylase and hexokinase activity in lactobacilli from traditionally prepared sourdoughs. Adv. Food Sci. 18:1-6. [Google Scholar]
- 59.Tanaka, K., et al. 2002. Two different pathways for D-xylose metabolism and the effect of xylose concentration on the yield coefficient of L-lactate in mixed-acid fermentation by the lactic acid bacterium Lactococcus lactis 10-1. Appl. Microbiol. Biotechnol. 60:160-167. [DOI] [PubMed] [Google Scholar]
- 60.Tanous, C., A. Gori, L. Rijnen, E. Chambellon, and M. Yvon. 2005. Pathways for α-ketoglutarate formation by Lactococcus lactis and their role in amino acid catabolism. Int. Dairy J. 15:759-770. [Google Scholar]
- 61.Titgemeyer, F., and W. Hillen. 2002. Global control of sugar metabolism: a Gram-positive solution. Antonie Van Leeuwenhoek 82:59-71. [PubMed] [Google Scholar]
- 62.Troyanskaya, O., et al. 2001. Missing value estimation methods for DNA microarrays. Bioinformatics 17:520-525. [DOI] [PubMed] [Google Scholar]
- 63.Van der Meulen, R., et al. 2007. Screening of lactic acid bacteria isolates from dairy and cereal products for exopolysaccharide production and genes involved. Int. J. Food Microbiol. 118:250-258. [DOI] [PubMed] [Google Scholar]
- 64.Van der Meulen, R., et al. 2007. Population dynamics and metabolite target analysis of lactic acid bacteria during laboratory fermentations of wheat and spelt sourdoughs. Appl. Environ. Microbiol. 73:4741-4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vermeulen, N., M. Pavlovic, M. A. Ehrmann, M. G. Gänzle, and R. F. Vogel. 2005. Functional characterization of the proteolytic system of Lactobacillus sanfranciscensis DSM 20451T during growth in sourdough. Appl. Environ. Microbiol. 71:6260-6266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vermeulen, N., M. Gänzle, and R. F. Vogel. 2006. Influence of peptide supply and cosubstrates on phenylalanine metabolism of Lactobacillus sanfransiscensis DSM20451T and Lactobacillus plantarum TMW1.468. J. Agric. Food Chem. 54:3832-3839. [DOI] [PubMed] [Google Scholar]
- 67.Vermeulen, N., M. G. Gänzle, and R. F. Vogel. 2007. Glutamine deamidation by cereal-associated lactic acid bacteria. J. Appl. Microbiol. 103:1197-1205. [DOI] [PubMed] [Google Scholar]
- 68.Vogelmann, S. A., M. Seitter, U. Singer, M. J. Brandt, and C. Hertel. 2009. Adaptability of lactic acid bacteria and yeasts to sourdough prepared from cereals, pseudo-cereals and cassava and use of competitive strains as starters. Int. J. Food Microbiol. 130:205-212. [DOI] [PubMed] [Google Scholar]
- 69.Vrancken, G., T. Rimaux, L. De Vuyst, and F. Leroy. 2008. Kinetic analysis of growth and sugar consumption by Lactobacillus fermentum IMDO 130101 reveals adaptation to the acidic sourdough ecosystem. Int. J. Food Microbiol. 128:58-66. [DOI] [PubMed] [Google Scholar]
- 70.Vrancken, G., T. Rimaux, S. Weckx, L. De Vuyst, and F. Leroy. 2009. Environmental pH determines citrulline and ornithine release through the arginine deiminase pathway in Lactobacillus fermentum IMDO 130101. Int. J. Food Microbiol. 135:216-222. [DOI] [PubMed] [Google Scholar]
- 71.Vrancken, G., T. Rimaux, D. Wouters, F. Leroy, and L. De Vuyst. 2009. The arginine deiminase pathway of Lactobacillus fermentum IMDO 130101 responds to growth under stress conditions of both temperature and salt. Food Microbiol. 26:720-727. [DOI] [PubMed] [Google Scholar]
- 72.Vrancken, G., et al. 2010. Yeast species composition differs between artisan bakery and spontaneous laboratory sourdoughs. FEMS Yeast Res. 10:471-481. [DOI] [PubMed] [Google Scholar]
- 73.Wang, H., K. A. Baldwin, D. J. O'Sullivan, and L. L. McKay. 2000. Identification of a gene cluster encoding Krebs cycle oxidative enzymes linked to the pyruvate carboxylase gene in Lactococcus lactis ssp. lactis C2. J. Dairy Sci. 83:1912-1918. [DOI] [PubMed] [Google Scholar]
- 74.Weckx, S., et al. 2009. Development and validation of a species-independent functional gene microarray that targets lactic acid bacteria. Appl. Environ. Microbiol. 75:6488-6495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Weckx, S., et al. 2010. Community dynamics of sourdough fermentations as revealed by their metatranscriptome. Appl. Environ. Microbiol. 76:5402-5408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Weckx, S., et al. 2010. Lactic acid bacteria community dynamics and metabolite production of rye sourdough fermentations share characteristics of wheat and spelt sourdough fermentations. Food Microbiol. 27:1000-1008. [DOI] [PubMed] [Google Scholar]
- 77.Wegmann, U., et al. 2009. Complete genome sequence of Lactobacillus johnsonii FI9785, a competitive exclusion agent against pathogens in poultry. J. Bacteriol. 191:7142-7143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Whiting, G. C., and R. A. Coggins. 1971. The role of quinate and shikimate in the metabolism of lactobacilli. Antonie Van Leeuwenhoek 37:33-49. [DOI] [PubMed] [Google Scholar]
- 79.Wolfinger, R. D., et al. 2001. Assessing gene significance from cDNA microarray expression data via mixed models. J. Comput. Biol. 8:625-637. [DOI] [PubMed] [Google Scholar]
- 80.Wood, B. J. B. 1998. Microbiology of fermented foods. Blackie Academic & Professional, London, United Kingdom.
- 81.Yvon, M., E. Chambellon, A. Bolotin, and F. Roudot-Algaron. 2000. Characterization and role of the branched-chain aminotransferase (BcaT) isolated from Lactococcus lactis subsp. cremoris NCDO 763. Appl. Environ. Microbiol. 66:571-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang, Z.-Y., et al. 2009. Complete genome sequence of Lactobacillus plantarum JDM1. J. Bacteriol. 191:5020-5021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhou, J. 2003. Microarrays for bacterial detection and microbial community analysis. Curr. Opin. Microbiol. 6:288-294. [DOI] [PubMed] [Google Scholar]