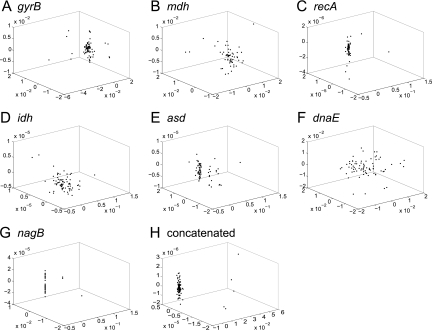
FIG. 2.
Nonmetric multidimensional scaling plots were constructed for pairwise distances between gene sequences from Vibrio cholerae isolates. Unique sequences are represented as closed circles, and the three-dimensional distances between coordinates approximate Hamming distances between sequences. The first seven plots represent sequences from individual loci, gyrB (A), mdh (B), recA (C), idh (D), asd (E), dnaE (F), and nagB (G); the last plot (H) displays distances for concatenated sequences from all MLSA loci.