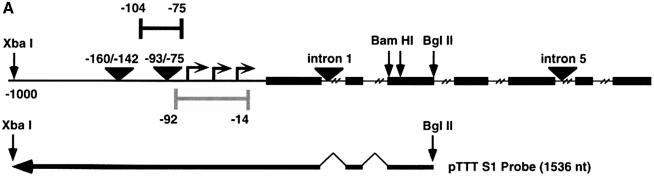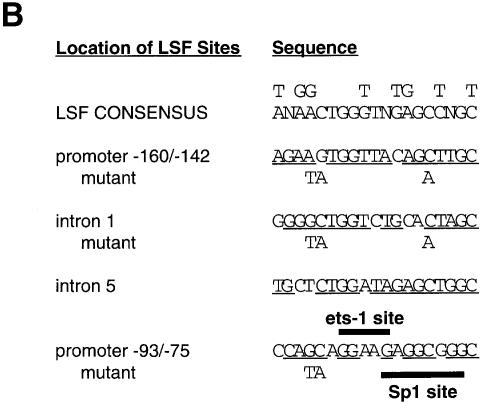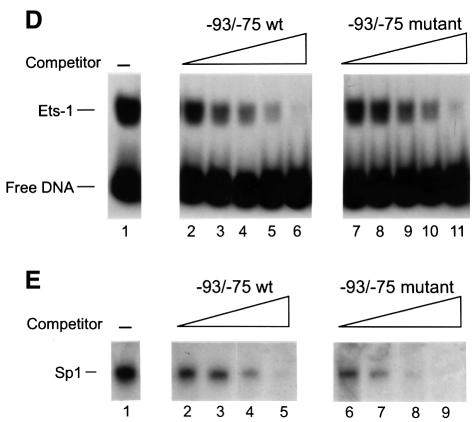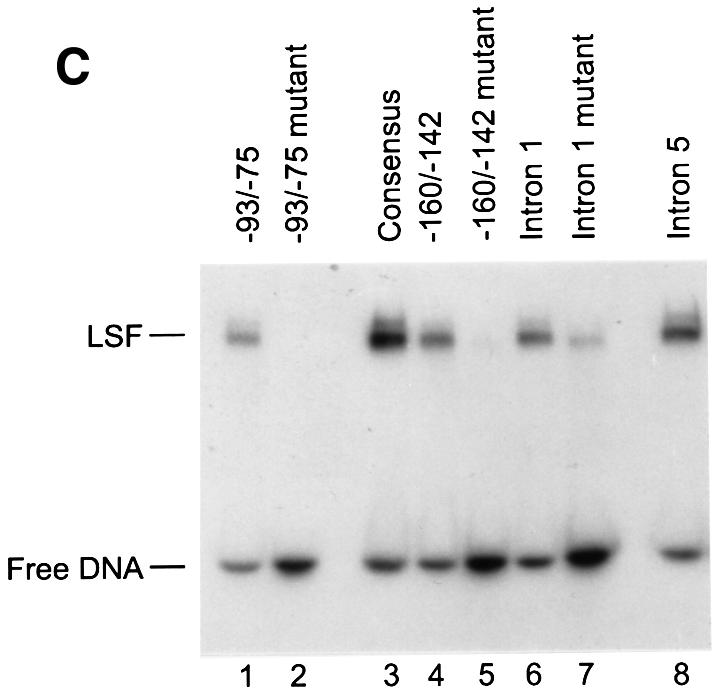
Fig. 1. LSF binds within regulatory regions of the mouse TS gene. (A) LSF sites located in promoter and intron regulatory regions of the mouse TS gene. Inverted triangles represent LSF binding sites; black boxes, coding regions; thick lines, promoter region; thin lines, introns; arrows, the positions of the multiple major transcriptional start sites (Deng et al., 1986); the gray bracket from –92 to –14, transcriptional initiation window; and the black bracket from –104 to –75, the essential promoter region (Geng and Johnson, 1993). Position +1 represents the A residue of the codon for translational initiation. The pTTT DNA probe used for S1 analysis (Ash et al., 1993) is indicated below. This probe extends from the BglII site to the XbaI site in the TS gene and lacks introns. The two BamHI sites mark the endpoints of the small 57 bp deletion in the minigenes that enables differentiation of the endogenous and minigene TS mRNAs in the S1 analysis. (B) Sequences of LSF wild-type binding sites in the mouse TS promoter and intron regulatory regions, and of the mutations generated in these sites. Underlined nucleotides in each site represent bases that match the LSF consensus binding site (Shirra, 1995). The locations of Ets-1 and Sp1 binding sites in the promoter –93/–75 region are indicated. The –160/–142 site is written in an inverted orientation relative to its position in the promoter in order to align the site with the consensus sequence. The base pair substitutions generated to mutate each site are indicated below the respective wild-type sequences. (C) EMSA of recombinant LSF with wild-type and mutant mouse TS sites, and the consensus LSF binding site, as indicated. Binding reactions contained either 400 ng of His-LSF (lanes 1 and 2) or 100 ng of His-LSF (lanes 3–8). (D) Mutant TS essential promoter region retains Ets-1 binding activity. Purified recombinant Ets-1 protein and the radiolabeled wild-type –93/–75 DNA were incubated with increasing amounts of wild-type (lanes 2–6) or mutant (lanes 7–11) competitor oligonucleotides, added at the same time as the radiolabeled DNA. The molar excess of competitor DNA was as follows: lanes 2 and 7, 2-fold; lanes 3 and 8, 5-fold; lanes 4 and 9, 10-fold; lanes 5 and 10, 20-fold; lanes 6 and 11, 50-fold. (E) Mutant TS essential promoter region retains Sp1 binding activity. Recombinant Sp1 and the radiolabeled wild-type –93/–75 DNA were incubated with increasing amounts of either wild-type (lanes 2–5) or mutant (lanes 6–9) competitors. The molar excess of competitor DNA was: lanes 2 and 6, 2-fold; lanes 3 and 7, 5-fold; lanes 4 and 8, 20-fold; lanes 5 and 9, 50-fold.