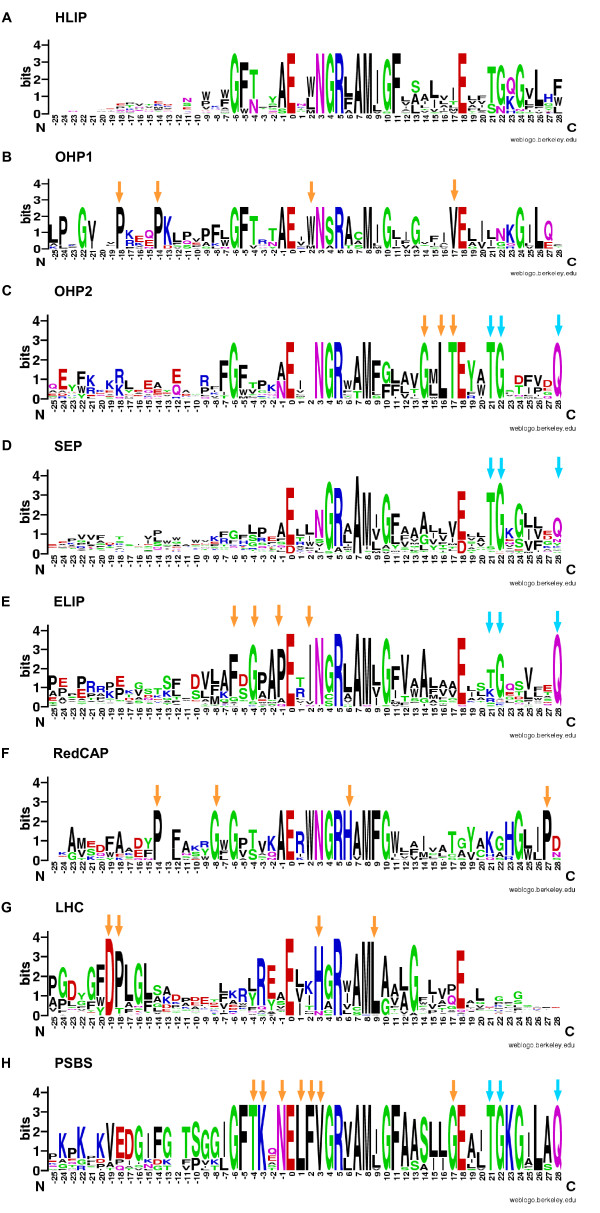
Figure 1.
Sequence logos of representative members of the extended LHC protein superfamily. (A-H) Sequence logos based on 54 amino acid positions surrounding the CB motif in the first helix were prepared from the same alignments that were used to build the HMM profiles. HMM analysis served as one of several criteria in the classification of unknown sequences (see Methods). The center of the x-axis corresponds to the highly conserved glutamate (E-0). For clarity, the sequence logo associated with the HMM profile of HLIPa is shown for HLIP and for the different SEP HMM profiles, the sequence logo of the combined SEP sequences is shown. An orange arrow marks amino acid residues that are specific to a particular family. Note that threonine (T-21), glycine (G-22) and glutamine (Q-28) are conserved in most SEPs, OHP2, ELIPs and PSBS (marked by a blue arrow). The new RedCAP family is described in more detail in the main text.