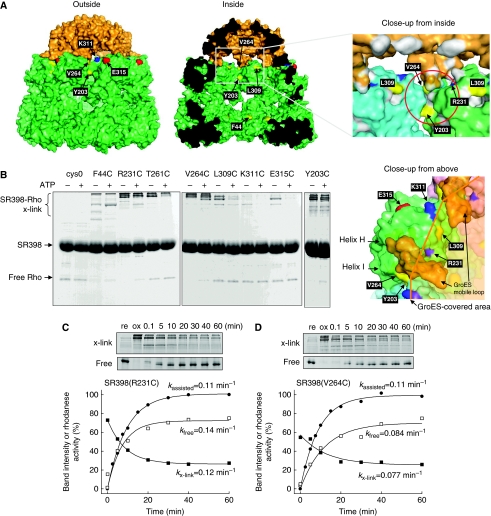
Figure 3.
GroEL residues that can interact with rhodanese in the cage. (A) Solvent-accessible surface of the GroES-associated GroEL ring in the GroEL–GroES–(ADP)7 complex (PDB code 1AON) viewed from outside, inside, close-up to the GroEL/GroES-binding site from inside and above. GroEL and GroES subunits are coloured in green and orange, respectively. The residues mutated to cysteine are labelled and coloured (hydrophilic, cyan; acidic, red; basic, blue; hydrophobic, yellow). The putative polypeptide-binding site is depicted by a red circle. (B) Cross-linking between rhodaneseHis6 (Rho) and cysteine-introduced SR398 mutants. Diamide was added to form disulphide bonds before (ATP−) or at 0.1 min after the addition of GroES and ATP (ATP+). Non-reducing SDS–PAGE gels were stained with CBB. (C, D) Time course of the cross-linking between rhodaneseHis6 and SR398(R231C) (C) or SR398(V264C) (D) during the assisted folding. After the folding reaction was started, aliquots were oxidized by diamide at indicated times. Time courses of cross-linked products (x-link, closed squares) and free rhodaneseHis6 (free, open squares), and rhodanese activity (closed circles) are plotted.