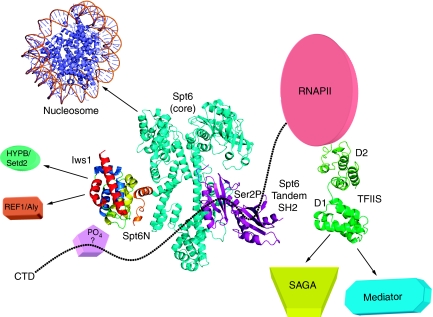
Figure 7.
Model of the transcription networks made by Iws1/Spt6 and TFIIS. Based on the results presented in this article as well as previously published data, the figure provides a current model of the Iws1/Spt6 complex and recapitulates our current knowledge of the transcription networks affected by Iws1/Spt6 and TFIIS, including homologous domains (ARM subdomain for Iws1 and Domain I for TFIIS). The model for Spt6 has been assembled from (i) the structure of the bacterial Tex protein that shares sequence homology with the core domain of Spt6 (cyan; Johnson et al, 2008), (ii) the recently discovered C-terminal tandem SH2 domains of Spt6 (purple; Diebold et al, 2010; Sun et al, 2010) and (iii) the Iws1/Spt6N complex (coloured as in previous figures; this manuscript). The N-terminal acidic domain of Spt6 is not included. Interactions of Iws1/Spt6 and TFIIS with different transcriptional effectors are shown with arrows. The C-terminal domain of the RNA polymerase II (CTD) is shown as dotted line. Binding of the Spt6 tandem SH2 repeats to Ser2-phosphorylated CTD repeats has been labelled with ‘Ser2P'. The putative CTD binding to the HEAT subdomain of Iws1, possibly through a phosphorylated residue and the requirement of an unknown factor, has been labelled with ‘?'. The PDB codes for the structures used to make the figure are 1AOI, 1WJT, 2XP1, 3BZC, 3GTM and 3GXW.