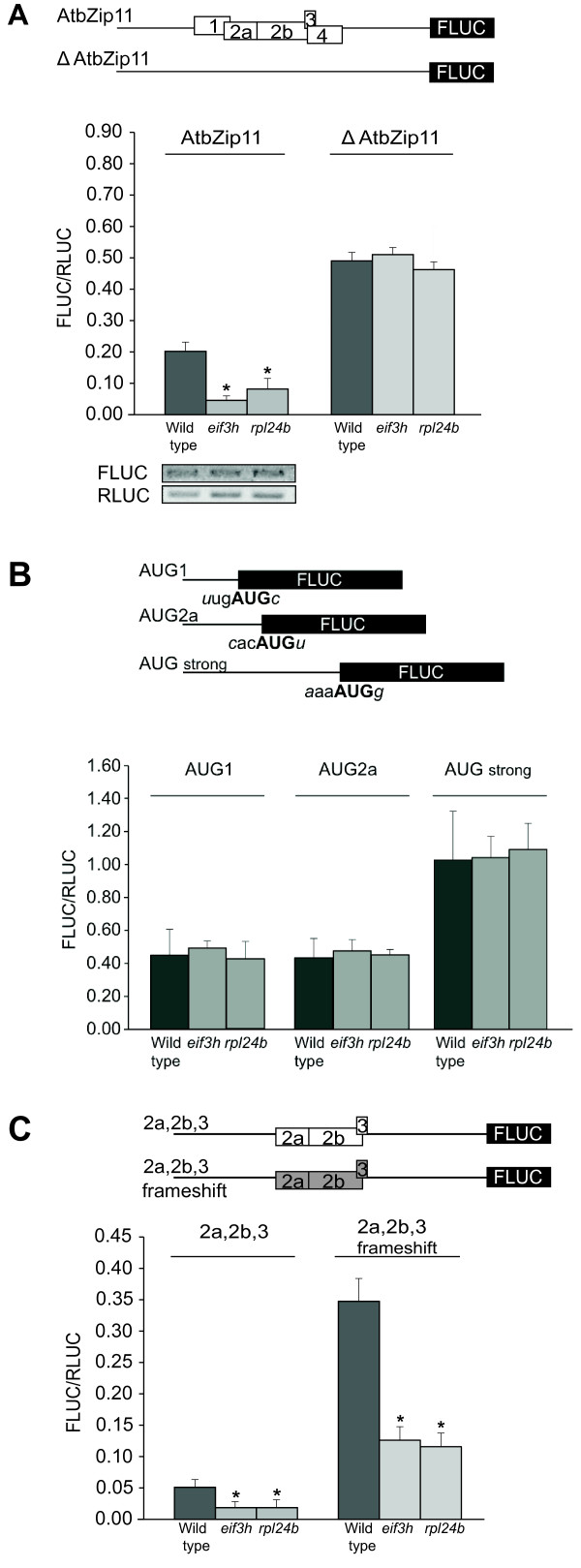
Figure 1.
Both eIF3h and RPL24B are required for efficient translation of the uORF-containing AtbZip11 mRNA. Data are from transient dual-luciferase gene expression assays in ten day old seedlings. The respective 5' leader is fused to firefly luciferase and is expressed in the presence of a reference gene expressing Renilla luciferase as an internal control for transformation efficiency. Both genes are transcribed from a CaMV 35S promoter [13]. (A) Schematic of the 5' leader of the AtbZip11 mRNA. Boxes numbered 1 to 4 represent uORFs. In the Δ AtbZip11 mutant the five uAUGs are replaced with stop codons [13]. Bars denote standard error; n = 7 to 10; * P < 0.002 by Student's t-test when compared to wild type. The gel images below the graph show mRNA levels for FLUC and RLUC as determined by subsaturating RT-PCR. Gel lanes correspond to the bars in the graph above. (B) Recognition of the uAUG start codons in AtbZip11. Like eif3h [13], rpl24b is not defective in recognizing the weak start codons, uAUG1 and uAUG2a, nor a strong version of uAUG4. (C) Testing the dependence of translation on the uORF peptide sequence. Both rpl24b and eif3h are inhibited to a similar degree by uORFs encoding the original uORF2 and uORF3 peptide sequences (white box in the schematic) and alternative peptide sequences generated by site-directed mutagenesis and a pair of compensatory frame shift mutations (gray box). For details on plasmid construction see [13].