Abstract
Chromatin-modifying complexes such as the NuRD complex are recruited to particular genomic sites by gene-specific nuclear factors. Overall, however, little is known about the molecular basis for these interactions. Here, we present the 1.9 Å resolution crystal structure of the NuRD subunit RbAp48 bound to the 15 N-terminal amino acids of the GATA-1 cofactor FOG-1. The FOG-1 peptide contacts a negatively charged binding pocket on top of the RbAp48 β-propeller that is distinct from the binding surface used by RpAp48 to contact histone H4. We further show that RbAp48 interacts with the NuRD subunit MTA-1 via a surface that is distinct from its FOG-binding pocket, providing a first glimpse into the way in which NuRD assembly facilitates interactions with cofactors. Our RbAp48·FOG-1 structure provides insight into the molecular determinants of FOG-1-dependent association with the NuRD complex and into the links between transcription regulation and nucleosome remodeling.
Keywords: Gene Regulation, Protein-Protein Interactions, Transcription Factors, Transcription Regulation, X-ray Crystallography, FOG-1, GATA-1, NuRD Complex, RbAp48
Introduction
Transcription repression and activation involve the coordinated recruitment and assembly of multiprotein complexes that remodel chromatin, increasing or decreasing the accessibility of specific sets of genes to the basal transcription machinery through one or more of several mechanisms. The NuRD (nucleosome remodeling and deacetylase) complex (also referred to as the Mi-2 complex) is one such complex that is conserved across multicellular plants and animals and is expressed in most or all tissues (1–4). NuRD is unique in that it contains both ATP-dependent chromatin-remodeling and histone deacetylase activities. The ATPase activity is conferred by CHD3 and CHD4 (chromodomain/helicase/DNA-binding protein; also known as Mi-2α and Mi-2β, respectively), whereas HDAC1 and HDAC2 are the catalytic histone deacetylase subunits.
The physical makeup of NuRD appears to vary in different cell types (5). CHD and HDAC proteins are invariably observed in NuRD purifications, and the retinoblastoma-associated proteins RbAp46 and/or RbAp48 (RBBP7 and RBBP4, respectively) are also core subunits of the complex. These two highly similar proteins (∼90% identical) were first identified through their interaction with the Rb tumor suppressor (6) and have been labeled as histone chaperones that function in the maintenance of chromatin structure. Despite their very high similarity, these two proteins appear to have at least partially distinct functions; for example, only RbAp48 has been detected in the chromatin assembly factor CAF-1 (7). Both RbAp46 and RbAp48 are found, however, with HDAC1/2 in several multiprotein complexes involved in the assembly, disassembly, and modification of chromatin structure, including NuRD (8, 9) and the Sin3 complex (10). Consistent with their roles in assembling or modifying chromatin structure, both RbAp46 and RbAp48 can recognize the N-terminal tail of histone H4 (11, 12).
Other core subunits of the NuRD complex include members of the MTA (metastasis-associated) and MBD (methyl-binding domain) protein families. Both MTA-1 and MTA-2 can inactivate p53-mediated apoptosis through the recruitment of deacetylase activity (13, 14), and MTA-1 is strongly linked to metastatic cell growth (15, 16). MTA proteins generally contain BAH (bromo-associated homology), ELM (Egl27 and MTA-1 homology), SANT (SWI3/ADA2/N-CoR/TFIIIB), and zinc finger domains, although for none of these domains has the function been clearly established. In general, MBD proteins recognize and bind to methylated CpG sequences in DNA (17). MBD2 and MBD3 are frequently found as part of NuRD (5), and in fact, the isoform of MBD3 that is most commonly associated with NuRD has an N-terminal deletion that most likely abrogates the ability of this protein to bind methylated DNA.
NuRD is essential for embryonic development in organisms ranging from nematodes to mammals (3). Mice lacking CHD4 display a number of T-cell developmental defects (18), and conditional deletion of Chd4 in bone marrow causes an accumulation of erythroid and lymphoid progenitors, with a loss of differentiated lymphoid and myeloid cells (19). Loss of RbAp46/48 in the specific context of NuRD has been shown to be associated with defects in chromatin structure correlated with aging (20), and as noted above, MTA-1 up-regulation is strongly correlated with cancer.
NuRD has been shown to regulate multiple genes during development (21). For example, during the early stages of hematopoietic stem cell differentiation, NuRD regulates the expression both of genes that are important during later stages and of genes that will become permanently silenced (19). In general, NuRD has been associated with gene repression, in line with both the presence of histone deacetylases in the complex and its association with a range of transcription repressors (1, 5). Perhaps surprisingly, however, NuRD has also been shown to function during transcription activation (22), and it is possible that NuRD complexes with different compositions exert different effects on gene targets.
Although the components of the NuRD complex have been identified in several cell types by biochemical purification and mass spectrometry (5), we still know relatively little about the functions of the individual subunits, the way they assemble, or the details of the mechanisms by which NuRD is recruited to target genes. It is known, however, that diverse nuclear factors associate with NuRD by binding to particular subunits. During hematopoietic development, for example, the transcription regulator FOG-1 (friend of GATA-1) is recruited to particular genomic loci by binding to the transcription factor GATA-1. GATA-1 is the master regulator of erythropoiesis, activating the expression of all known erythroid and megakaryocytic genes (23) and also repressing genes such as the hematopoietic stem cell factor Gata2 (24). The GATA-1/FOG-1 interaction is required for most GATA-1 functions (25, 26), but the mechanism by which this complex results in alterations in the expression of target genes is only partially understood. We showed previously that FOG-1 can bind to the NuRD complex and that the interaction is mediated by the 15 N-terminal amino acids of FOG-1 (22, 27). This motif is both necessary and sufficient for high affinity NuRD binding in vitro and in vivo and for mediating both transcription activation and repression by FOG-1. However, the molecular basis of this interaction, or indeed any interaction between NuRD and a transcription regulator, has remained unresolved. Here, we present the x-ray crystal structure of the complex formed between RbAp48 and FOG-1-(1–15). We also probe the interplay between FOG-1, RbAp48, and MTA-1, demonstrating that the FOG-1- and MTA-1-binding sites on RbAp48 are separable. These data provide the first molecular details of the mechanism through which the NuRD complex associates with gene-specific transcription regulators.
EXPERIMENTAL PROCEDURES
Peptide Synthesis
The FOG-1-(1–12), FOG-1-(1–15), and biotinylated FOG-1-(1–15) peptides were chemically synthesized and purified by Auspep (Tullamarine, Australia).
Cloning, Expression, and Purification
Full-length RbAp48 (residues 1–425; UniProt accession number Q09028) was cloned into a pFBDM vector encoding an N-terminal His6 tag and a thrombin protease cleavage site. Recombinant baculovirus was generated in Sf9 cells using the Bac-to-Bac (Invitrogen) expression methodology. Briefly, cells were transfected for 72 h to yield virus containing the RbAp48 gene. The recombinant primary virus was subsequently amplified to yield high titer secondary virus used for protein expression. Sf9 cells cultured in SF-900 II SFM medium (Invitrogen) and infected with the recombinant virus were harvested 80 h post-infection by pelleting at 4000 rpm for 20 min at 4 °C. Cell pellets were stored in −80 °C until required.
Cells from 1 liter of expression culture were resuspended on ice in 30 ml of lysis buffer consisting of 20 mm Tris (pH 8.0), 150 mm NaCl, 10 mm MgCl2, and 1 mm β-mercaptoethanol supplemented with EDTA-free protease inhibitors (Roche Applied Science). Lysis was performed by passing the cell suspension through an EmulsiFlex C5 instrument (Avestin). Nonidet P-40 (0.1%, v/v) and DNase I were added to the lysate before cell debris was pelleted at 20,000 rpm for 40 min at 4 °C. The supernatant was transferred to 2.5 ml of nickel-nitrilotriacetic acid Superflow beads (Qiagen) and rotated for 2 h at 8 °C to facilitate protein binding. Beads were then pelleted for 10 min at 2400 × g and transferred to a polypropylene gravity flow column (Bio-Rad). The beads were first washed with 20 column volumes of 20 mm Tris (pH 8.0), 300 mm NaCl, and 2 mm imidazole and then in the same buffer containing 150 mm NaCl and stepwise increasing concentrations of imidazole up to 40 mm before elution with 0.5 m imidazole into 5-ml fractions. Fractions containing RbAp48 were identified by SDS-PAGE, pooled, and dialyzed overnight using an 8000-Da molecular mass cutoff membrane in 2 liters of dialysis buffer containing 20 mm Tris (pH 8.0), 150 mm NaCl, and 2.5 mm CaCl2. The hexahistidine affinity tag was cleaved from the protein by adding 4 μl of thrombin protease (1.7 units/μl; Novagen) to the dialysate.
Following dialysis, RbAp48 was concentrated with a Vivaspin 10,000-Da molecular mass cutoff filter to a volume of 5–8 ml and diluted in 20 mm Tris (pH 7.5) to bring the final NaCl concentration down to 30–35 mm. DTT and Nonidet P-40 were added to the protein solution to final concentrations of 10 mm and 0.02% (v/v), respectively. The sample was then applied to a Mono Q HR 5/5 column (1-ml column volume), and unbound protein was washed off with 20 mm Tris (pH 7.5). Bound protein was eluted from the column using a 0–50% gradient of 20 mm Tris (pH 7.5), and 1 m NaCl over 80 ml. Fractions containing RbAp48 were identified by SDS-PAGE, pooled, and buffer-exchanged into 20 mm Tris (pH 7.5) and 100 mm NaCl before concentration in a Vivaspin 10,000-Da molecular mass cutoff filter to a final concentration of 8 mg/ml.
Isothermal Titration Calorimetry
Purified RbAp48 was dialyzed against buffer containing 20 mm Tris (pH 7.5), 150 mm NaCl, and 1 mm DTT. The FOG-1-(1–12) peptide was dissolved in the same buffer. The FOG-1 peptide (100 μm) was titrated into RbAp48 (10 μm, 200 μl) in a series of 26 1.5-μl injections, with a 5-min interval between each injection, using a MicroCal iTC200 titration calorimeter. The reference power was set at 2 μcal/s, and the cell was stirred continuously at 1000 rpm. The evolved heats were integrated and normalized for protein concentration. After base-line correction (using data from titration of the FOG-1 peptide into buffer), the data were fitted using a single-site model and MicroCal Origin 5.0.
Surface Plasmon Resonance
Biotinylated FOG-1-(1–15) was immobilized onto a streptavidin-coated Biacore SA sensor chip to a level of 120 resonance units. Increasing concentrations of RbAp48 were injected over the chip at a flow rate of 20 μl/min in PBS containing 10 mm phosphate (pH 7.4), 150 mm NaCl, 1 mm DTT, and 0.01% surfactant P20. Binding events were visualized as a positive change in resonance units and recorded as a function of time. Data analysis was performed with BIAevaluation software (Biacore AB).
Crystallization and Structure Determination
Synthetic FOG-1-(1–15) peptide was dissolved in demineralized water at a concentration of 1.1 μm. The peptide solution was then added to purified and concentrated RbAp48 to obtain a molar protein/peptide ratio of 1:1.2. Crystallization trials were set up in 96-well plates as sitting drops using 0.2 μl each of protein solution and crystallization solution. Crystals were obtained after incubation overnight in 4 °C in several conditions in the Index HT (Hampton Research) screen. After optimization and microseeding trials using an Oryx6 protein crystallization robot (Douglas Instruments), single crystals were obtained in condition 87 (20% PEG 3350 and 0.2 m sodium malonate (pH 7.0)). These crystals were harvested into a cryoprotectant solution containing 25% PEG 400 in mother liquor before cryocooling in liquid nitrogen. X-ray diffraction data were collected on beamline I03 at the Diamond Light Source (Oxfordshire, United Kingdom). Data were processed and scaled using MOSFLM (28) and Scala (29). The structure of the complex was phased using the molecular replacement function in the program Phaser (30). Clear solutions for two molecules in the asymmetric unit were found using the unbound structure of RbAp48 as a search model (Protein Data Bank code). Manual model building and crystallographic refinement using Coot (31) and Refmac (32) allowed completion of the structure to a resolution of 1.9 Å. Validation of the structure was carried out with MolProbity (33). Interface analysis was performed using the EBI PISA server (34), and figures were generated using PyMOL (35). The coordinates and structure factors (code 2XU7) have been deposited in the Protein Data Bank.
Pulldown Assays
RbAp48 mutants were designed based on visual inspection of the RbAp48·FOG-1 crystal structure. The double mutant E126A/E179A (residues that contact Lys-5 of FOG-1) and the quadruple-point mutant E231A/D248A/N277A/E319A (residues that contact Arg-3 and Arg-4 of FOG-1) were generated using the QuikChange multisite-directed mutagenesis kit (Stratagene). Wild-type and mutant RbAp48 and MTA-1 were in vitro translated using the TnT quick coupled transcription/translation system (Promega). For the pulldown assays, ∼10 μg of biotinylated FOG-1-(1–15) peptide was immobilized onto high capacity streptavidin beads, which were then resuspended in 25× bead volume of binding buffer (Tris (pH 7.4), 300 mm NaCl, 0.5% IGEPAL, 1× Complete EDTA-free protease inhibitor (Roche Applied Science)). Rabbit reticulocyte lysate containing in vitro translated 35S-labeled RbAp48 (15 μl) was incubated with a suspension (250 μl) of biotinylated FOG-1-(1–15)-bound streptavidin beads for 1 h at 4 °C. For negative control pulldown assays, streptavidin beads alone were used instead. Beads were washed five times with binding buffer. Proteins from the pulldown assays were analyzed by SDS-PAGE and then visualized by autoradiography. The pulldown experiment showing the interaction between FOG-1-(1–15), MTA-1, and E126A/E179A mutant RbAp48 was carried out as described above except that 45 μl of reticulocyte lysate containing 35S-labeled MTA-1 was also added during the incubation. For GST pulldown assays to test the RbAp48/H4 interaction, GST-H4-(1–48) was expressed and purified by glutathione affinity chromatography as described previously (11). Pulldown experiments with 35S-labeled RbAp48 were then carried out as described above.
RESULTS
FOG-1 Binds to Purified Recombinant RbAp48
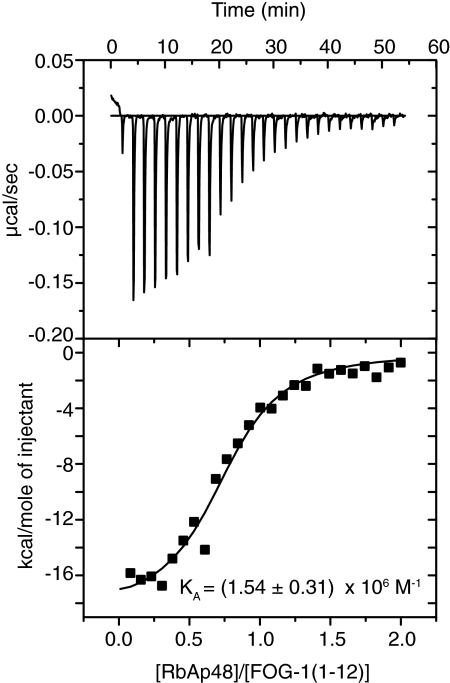
We previously reported that FOG-1-(1–15) can interact with RbAp48 in the context of GST pulldown assays carried out using RbAp48 expressed in a reticulocyte lysate (27). To confirm that this interaction is direct, we first expressed RbAp48 with an N-terminal His tag in Sf9 insect cells using the Bac-to-Bac expression system and then purified the protein using nickel affinity and ion exchange chromatography. To determine the binding affinity of RbAp48 for FOG-1, isothermal titration calorimetry was carried out. Fig. 1 (upper panel) shows the heat evolved when FOG-1-(1–12) was titrated into RbAp48. The heat peaks were integrated, and the binding affinity was calculated to be (1.54 ± 0.31) × 106 m−1 (Fig. 1, lower panel). A similar affinity ((1.36 ± 0.07) × 106 m−1, χ2 = 10.8) was determined by surface plasmon resonance experiments in which FOG-1-(1–15) bearing an N-terminal biotin tag was immobilized onto a streptavidin-coated surface plasmon resonance chip and treated with increasing concentrations of purified RbAp48 (supplemental Fig. 1).
FIGURE 1.
Determination of the affinity of RbAp48 for FOG-1-(1–12) using isothermal titration calorimetry. Raw (upper panel) and integrated (lower panel) data are shown from a titration of FOG-1-(1–12) into RbAp48. The fit to a simple 1:1 binding model is also shown.
Determination of the RbAp48·FOG-1-(1–15) Structure
We next reconstituted the RbAp48·FOG-1 complex by combining purified full-length RbAp48 with a synthetic peptide comprising FOG-1-(1–15) and screened for solution conditions under which the complex would crystallize. We observed crystals after 1–2 days in 4 °C; these crystals grew in a monoclinic space group with two copies of the complex per asymmetric unit. After optimization of protein concentration and improving crystal morphology through the use of microseed matrix screening (36), the crystals diffracted to 1.9 Å resolution, and data were collected at the Diamond Light Source. The structure of the complex was solved by molecular replacement using the structure of RbAp48 as an initial search model and refined to an R factor of 18.5% and an Rfree factor of 22.5%. Data collection and refinement statistics are outlined in Table 1. The final model consists of RbAp48 residues 11–410 and FOG-1 residues 1–13. The remainder of the protein residues (positions 1–10 and 411–425) could not be modeled due to poor electron density, probably due to motional disorder in the crystal. The two copies of RbAp48 in the asymmetric unit are highly similar and superimpose with a root mean square deviation of 0.52 Å over 355 Cα atoms. We note that there are interactions near the FOG-1-binding site both within the asymmetric unit and between symmetry-related RbAp48 molecules, but no direct contacts are made between the RbAp48 chain and a FOG-1 peptide belonging to a separate complex.
TABLE 1.
Data collection and refinement statistics
Values for the highest resolution shell are shown in parentheses. r.m.s.d., root mean square deviation.
| Data collection | P21 |
| Cell dimensions | |
| a, b, c (Å) | 75.75, 59.84, 100.65 |
| α, β, γ | 90°, 93.55°, 90° |
| Resolution (Å) | 100.5-1.9 (2.0-1.9) |
| Rmerge (%) | 8.0 (44.7) |
| I/σ(I) | 11.3 (3.1) |
| Completeness (%) | 99.5 (99.9) |
| Redundancy | 3.2 (3.2) |
| Refinement | |
| Resolution (Å) | 100.5-1.9 (1.949-1.9) |
| No. of reflections | 67,143 (4936) |
| Rwork/Rfree | 0.18/0.22 |
| No. of atoms | |
| Protein | 6172 |
| Ligand | 25 |
| Water | 225 |
| B-factors | |
| Protein | 12.5 |
| Ligand | 30.3 |
| Water | 15.5 |
| r.m.s.d. values | |
| Bond length (Å) | 0.023 |
| Bond angle | 1.86° |
| Ramachandran values | |
| Most favored (%) | 89.8 |
| Additional allowed (%) | 10.2 |
| Disallowed (%) | 0.0 |
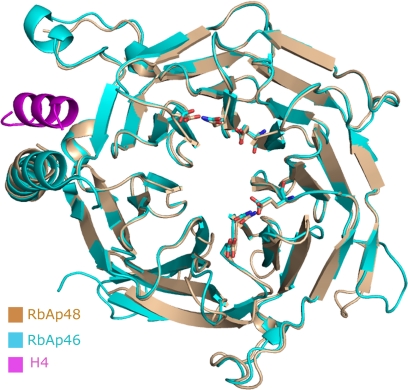
FOG-1 Recognizes One Face of the Central Cavity of RbAp48
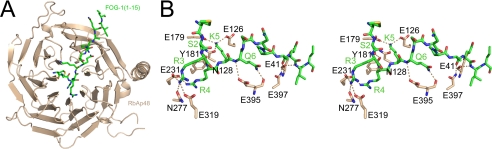
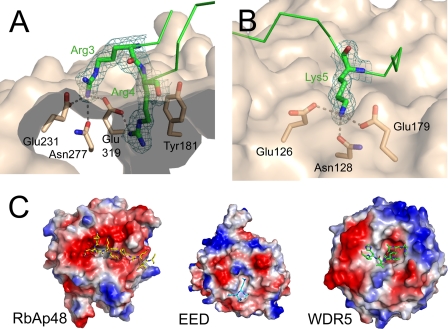
RbAp48 folds into a seven-bladed β-propeller structure with a prominent N-terminal α-helix (Fig. 2). Several loop regions could be traced that are disordered in the unbound RbAp48 structure (Protein Data Bank code 3GFC), which is currently unpublished, but overall, the protein conformation is not significantly affected by formation of the complex with FOG-1 (root mean square deviation of 0.45 Å over 329 Cα atoms). A superposition of the RbAp48·FOG-1 structure onto the structure of RbAp46 bound to an N-terminal peptide from histone H4 (Protein Data Bank code 3CFV) (11) similarly shows that the conformations of the two RbAp proteins are very similar, as expected given their high sequence identity. In particular, the putative histone H4-binding region in RbAp48 (by homology with RbAp46 (11)) closely resembles its counterparts in RbAp46 and Drosophila p55 (37). Surprisingly, however, FOG-1 does not contact the H4-binding site but instead binds to RbAp48 in a largely extended conformation across the axial channel on the “top” surface of the β-propeller (i.e. the smaller of the two flat axial surfaces). The RbAp48/FOG-1 interaction is highly specific, with a total of 8 of 13 FOG-1 residues participating in hydrogen bond or ion pair contacts with RbAp48 (Fig. 2 and Table 2). The binding pocket is lined with a large number of aspartate and glutamate residues, which are situated in loops between the RbAp48 propeller blades and together create a highly negatively charged surface that accommodates the positively charged FOG-1 residues Arg-3, Arg-4, and Lys-5 (Fig. 2). The side chain of Arg-4 extends directly into the axial channel, where it forms specific electrostatic and hydrogen bond interactions with RbAp48 residue Glu-231, and its guanidino group is in a favorable orientation to make cation-π interactions with Tyr-181 (Fig. 3). In neighboring pockets, Arg-3 forms a hydrogen bond with Asn-277 and also ion pairs with Glu-231 and Glu-319, whereas Lys-5 forms polar interactions with both Glu-179 and Glu-126 and a hydrogen bond with Asn-128 (Fig. 3). Additional specific interactions include several hydrogen bonds between Gln-6 and Glu-395, Ser-2 and Tyr-181, and Gln-11 and Glu-41. Further stabilization of the interaction is provided by RbAp48 residues Lys-376 and Asn-397, which interact with the main chain carbonyl groups of Arg-4 and Arg-10, respectively. It is notable that the locations and identities of all of the FOG-1-interacting residues in RbAp48 are conserved in RbAp46 (Fig. 4), consistent with our previous data showing that FOG-1 can bind to both RbAp46 and RbAp48 in pulldown assays (27). The specificity of the interaction is further underscored by the observation that RbAp48 does not bind measurably to an N-terminal peptide derived from MBD3, another NuRD component (data not shown). This sequence (MERKRWECPALPQGW) contains a triple basic RKR sequence, demonstrating that such an array of positively charged side chains alone is not sufficient to specify binding to RbAp48.
FIGURE 2.
A, view of FOG-1-(1–15) (green) bound to the top surface of RbAp48 (beige). B, stereo image of the FOG-1-(1–15) peptide-binding site. Interactions between FOG-1 residues (green) and RbAp48 residues (beige) are indicated as dotted lines. Details of the interactions are given in Table 2.
TABLE 2.
Specific interactions at the RbAp48/FOG-1 interface
| FOG-1 residue | RbAp48 residue | Interaction type and distance |
|
|---|---|---|---|
| H-bond | Salt bridge | ||
| Å | |||
| Ser-2 | Tyr-181 | 2.4 | |
| Arg-3 | Glu-319 | 3.0 | 3.3 |
| Arg-3 | Asn-277 | 2.9 | |
| Arg-3 | Glu-231 | 3.1 | 3.1 |
| Arg-4 | Asn-277 | 3.4 | |
| Arg-4 | Glu-231 | 2.8 | 2.8 |
| Lys-5 | Asn-128 | 2.7 | |
| Lys-5 | Glu-179 | 2.7 | 2.7 |
| Lys-5 | Glu-126 | 2.8 | 2.8 |
| Gln-6 | Glu-395 | 3.8 | |
| Gln-6 | Glu-395 | 2.8 | |
| Arg-10 | Asn-397 | 2.9 | |
| Gln-11 | Glu-41 | 2.5 | |
| Ile-12 | Glu-41 | 2.7 | |
FIGURE 3.
A and B, close-up views of the Arg-3 and Arg-4 (A) and Lys-5 (B) interactions with RbAp48. Hydrogen bonds are indicated by dotted lines. The molecular surface of RbAp48 is shown in beige, and FOG-1 is shown as green sticks, with the 2mFo − DFc omit electron density map contoured at 0.30 electrons/Å3 (1.0σ) covering the FOG-1 residues. C, comparison of electrostatic surface potentials of the binding surfaces of RbAp48, EED, and WDR5. The proteins are shown as molecular surfaces colored according to electrostatic potential, and bound peptides are shown in stick representation. Left panel, RbAp48 in complex with FOG-1-(1–15) (shown as yellow sticks). The high proportion of negatively charged side chains (red) is notable. Middle panel, EED in complex with a H3K27me peptide (Protein Data Bank code 3JZG). Right panel, WDR5 in complex with a histone H3 peptide (code 3EMH).
FIGURE 4.
Structural alignment of RbAp48 (beige) and RbAp46 (cyan). FOG-1-binding residues in RbAp48 and the corresponding residues in RbAp46 are shown as sticks. The histone H4 peptide bound to RbAp46 (Protein Data Bank code 3CFS) is shown in magenta. The root mean square deviation for superposing 349 Cα atoms is 0.69 Å.
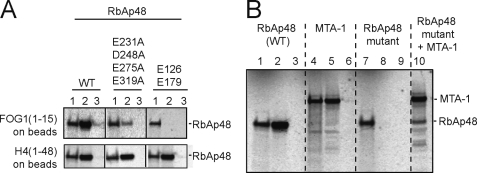
To validate our structural data, we carried out site-directed mutagenesis of RbAp48, targeting residues predicted to interact with FOG-1-(1–15) (Fig. 5A). Simultaneous mutation of Glu-126 and Glu-179, which both recognize Lys-5, impairs the interaction, as does the quadruple mutation E231A/D248A/E275A/E219A (all residues that lie in the vicinity of Arg-3 and Arg-4). To confirm that the RbAp48 mutants are all correctly folded, we demonstrated that they can both bind to histone H4.
FIGURE 5.
A, pulldown experiments showing that either a double-point mutation of Lys-5-binding residues (Glu-126, Glu-179) or a quadruple-point mutation of RbAp48 residues near Arg-3/Arg-4 (Glu-231, Asp-248, Asn-277, Glu-319) disrupts binding to FOG-1. The integrity of both of these mutants is demonstrated by the retention of their ability to bind to histone H4 (11). For each interaction, lane 1 represents 10% input protein, lane 2 shows the pulldown of in vitro translated 35S-labeled RbAp48 protein using biotinylated FOG-1-(1–15) or GST-H4-(1–48) as bait, and lane 3 is a control in which no biotinylated FOG-1-(1–15) was added to the streptavidin beads or GST alone was added to glutathione beads. B, pulldown experiments showing the interaction between FOG-1-(1–15), MTA-1, and RbAp48. Lanes 1, 4, and 7 show loading controls for the indicated protein (10% of input). Lanes 2, 5, 8, and 10 show the pulldown of each protein by biotinylated FOG-1-(1–15) immobilized on streptavidin beads. Lanes 3, 6, and 9 show the pulldown of each protein by streptavidin beads alone. The RbAp48 mutant is E126A/E179A. Lane 10 shows that RbAp48 and FOG-1 are not competing for the same binding site on MTA-1.
RbAp48 Binds FOG-1 and MTA-1 Independently
It is noteworthy that neither FOG-1 nor the other proteins containing the RbAp48-binding motif have been demonstrated to recruit other complexes containing RbAp48 and RbAp46, such as the Sin3 and PRC2 complexes (27). The question therefore arises as to how this type of specificity is generated, given that the different complexes share several subunits. It is possible that NuRD-specific proteins play a role in the generation of such specificity. We have shown previously that the NuRD component MTA-1, which is not present in the Sin3 and PRC2 complexes, interacts with FOG-1 (27), and it is possible that multiple interactions with both RbAp48 (and/or RbAp46) and MTA-1 are required for efficient recruitment of NuRD to FOG-1-dependent promoters. In fact, both Arg-4 and Lys-5 of FOG-1 are also essential for the FOG-1/MTA-1 interaction (27). However, given that these residues are mostly buried in the RbAp48·FOG-1 complex, it is unlikely that both MTA-1 and RbAp48 could be interacting simultaneously with the same molecule of FOG-1. Taken together, these data suggest that RbAp48 and MTA-1 likely bind to the same motif in different molecules of FOG-1. We used pulldown experiments to test this hypothesis. As expected, both MTA-1 and wild-type RbAp48 bound to immobilized FOG-1-(1–15) (Fig. 5B, lanes 2 and 5), whereas E126A/E179A mutant RbAp48 did not (lane 8). Importantly, we found that the RbAp48 mutant could be recruited by FOG-1-(1–15) in the presence of MTA-1 (Fig. 5B, lane 10) by virtue of FOG-1/MTA-1 and MTA-1/RbAp48 interactions. Thus, these data demonstrate that the FOG-1 and MTA-1 recognition sites on RbAp48 are distinct and separable and also that the FOG-1- and RbAp48-binding sites on MTA-1 are likewise separable.
DISCUSSION
Conserved Residues in FOG-like Motifs
A sequence alignment of human proteins known to contain the N-terminal RRKQXXP motif is shown in Fig. 6. The general conservation of this motif in several co-repressors and transcription factors, including the transcription factor SALL1 (38), EBFAZ (early B-cell factor-associated zinc finger protein), and the B-cell leukemia/lymphoma proteins BC11A and BC11B (also known as CTIP1 and CTIP2 (C-terminal binding protein-interacting protein), respectively), suggests that they are likely to recruit the NuRD complex in a similar manner. The structure of FOG-1-(1–15) bound to RbAp48 described here underscores the dependence of the binding on the RRKQ submotif, as the majority of the specific interactions are conferred by these residues. In particular, FOG-1 residues Arg-3, Arg-4, and Lys-5 have been found to be important for the interaction, as mutation in any one of these positions results in the loss of NuRD binding and GATA-1-mediated repression (27, 39). Consistent with these data, the side chains of the two non-conserved residues immediately following the RRKQ submotif (Ser and Asn in FOG-1) are not engaged in interactions with RbAp48 but are instead oriented toward the solvent.
FIGURE 6.
Sequence alignment of the 12 N-terminal residues of human FOG-1, FOG-2, CTIP1 (BC11A), CTIP2 (BC11B), and SALL1 and murine EBFAZ. The consensus sequence is shown below the alignment.
Differences in Binding Modes between RbAp48 and Other WD40 Proteins
The structures of several other WD40 proteins, including the transcription regulator WDR5 (WD repeat protein 5) and the Polycomb protein EED in complexes with binding partners, show that interactions of these proteins also involve the top of the β-propeller. Structures of WDR5 bound to methylated peptides from either histone H3 or MLL1 (mixed lineage leukemia protein 1) (40–43) all show the axial channel accepting an arginine residue from the binding partner (Arg-2 in H3 and Arg-3765 in MLL1). In contrast, structures of EED bound to methylated histone H3 peptides reveal that EED lacks the axial channel altogether, instead accommodating a methylated lysine in a pocket on its surface. The EED and WDR5 complex structures also lack the large number of highly specific intermolecular interactions that are observed in and adjacent to the FOG-1-binding channel in RbAp48. Furthermore, a comparison of the electrostatic surfaces of the three proteins reveals a much less charged interaction surface on top of the EED and WDR5 β-propellers compared with RbAp48 (Fig. 3).
Interplay between FOG-1, RbAp48, and MTA-1
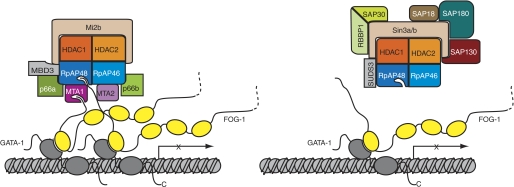
Our data show that each of the three proteins MTA-1, RbAp48, and FOG-1 can form pairwise interactions with the other two and that, in the case of FOG-1, it is the same motif that binds to the two different partners. In wild-type NuRD in vivo, it is possible that RbAp46, RbAp48, and MTA-1 can simultaneously contact separate FOG-1 molecules. These data are consistent with a model in which recruitment of NuRD is optimal when multiple FOG-1 molecules are present at the same locus and several points of contact can be made (Fig. 7). In this model, the recruitment of Sin3 or PRC2, which both lack MTA-1, would be less likely. In this context, it is notable that many promoters contain multiple binding sites for relevant transcription factors and that many transient transfection assays are carried out using artificial promoter sequences that contain multiple recognition sites for the transcription factor under investigation (44). It is possible that such arrangements are functionally superior because of their improved ability to recruit co-regulator complexes such as NuRD in a multivalent manner. Overall, our data underscore the role of RbAp48 as a versatile component of protein complexes involved in regulation of chromatin structure.
FIGURE 7.
Schematic model indicating the possible origin of the specificity of FOG-1 for NuRD (rather than Sin3). Zinc fingers of FOG-1 are shown as yellow ellipses, and zinc fingers of GATA-1 (which is bound to a GATA site at a gene promoter) are shown as gray ellipses. It is possible that recruitment of NuRD requires multiple points of interaction (left panel). This situation would be favored at promoters containing multiple GATA sites, which could have multiple FOG-1 molecules associated with them. The Sin3 complex lacks MTA subunits and might therefore have lower avidity for FOG-1-rich promoter regions (right panel).
In summary, we have demonstrated that FOG-1 binds to NuRD via a mechanism involving a highly specific interaction of its N terminus with a novel binding pocket in RbAp48. It is likely that a similar mechanism is used by other transcription cofactors to recruit NuRD via a FOG-1-like motif. The results suggest that multiple interactions with this and other NuRD components such as MTA-1 are likely to contribute to the recruitment of NuRD to genomic sites in vivo and perhaps explain the specificity of FOG-1 for NuRD in comparison with the related Sin3 and PRC2 complexes. Our structure provides a basis for further experiments to resolve the structural determinants underlying the assembly, regulation, and promoter targeting of NuRD.
Supplementary Material
Acknowledgments
We thank the staff at the Diamond Light Source for beam time and help with data collection, Philippa Stokes for assistance with isothermal titration calorimetry, and Stephen McLaughlin for assistance with surface plasmon resonance experiments.
Footnotes
This work was supported in part by National Institutes of Health Grant DK058044. This work was also supported by grants from the Australian National Health and Medical Research Council and the Wellcome Trust, United Kingdom.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
The atomic coordinates and structure factors (code 2XU7) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
REFERENCES
- 1. Denslow S. A., Wade P. A. (2007) Oncogene 26, 5433–5438 [DOI] [PubMed] [Google Scholar]
- 2. Xue Y., Wong J., Moreno G. T., Young M. K., Côté J., Wang W. (1998) Mol. Cell 2, 851–861 [DOI] [PubMed] [Google Scholar]
- 3. Ahringer J. (2000) Trends Genet. 16, 351–356 [DOI] [PubMed] [Google Scholar]
- 4. Gregory G., Miccio A., Bersenev A., Wang Y., Hong W., Zhang Z., Poncz M., Tong W., Blobel G. A. (2010) Blood 115, 2156–2166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bowen N. J., Fujita N., Kajita M., Wade P. A. (2004) Biochim. Biophys. Acta 1677, 52–57 [DOI] [PubMed] [Google Scholar]
- 6. Qian Y. W., Wang Y. C., Hollingsworth R. E., Jr., Jones D., Ling N., Lee E. Y. (1993) Nature 364, 648–652 [DOI] [PubMed] [Google Scholar]
- 7. Verreault A., Kaufman P. D., Kobayashi R., Stillman B. (1996) Cell 87, 95–104 [DOI] [PubMed] [Google Scholar]
- 8. Henikoff S. (2003) Nature 423, 814–817 [DOI] [PubMed] [Google Scholar]
- 9. Loyola A., Almouzni G. (2004) Biochim. Biophys. Acta 1677, 3–11 [DOI] [PubMed] [Google Scholar]
- 10. Silverstein R. A., Ekwall K. (2005) Curr. Genet. 47, 1–17 [DOI] [PubMed] [Google Scholar]
- 11. Murzina N. V., Pei X. Y., Zhang W., Sparkes M., Vicente-Garcia J., Pratap J. V., McLaughlin S. H., Ben-Shahar T. R., Verreault A., Luisi B. F., Laue E. D. (2008) Structure 16, 1077–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Verreault A., Kaufman P. D., Kobayashi R., Stillman B. (1998) Curr. Biol. 8, 96–108 [DOI] [PubMed] [Google Scholar]
- 13. Luo J., Su F., Chen D., Shiloh A., Gu W. (2000) Nature 408, 377–381 [DOI] [PubMed] [Google Scholar]
- 14. Moon H. E., Cheon H., Lee M. S. (2007) Oncol. Rep. 18, 1311–1314 [PubMed] [Google Scholar]
- 15. Pencil S., Toh Y., Nicolson G. (1993) Breast Cancer Res. Treat. 25, 165–174 [DOI] [PubMed] [Google Scholar]
- 16. Toh Y., Pencil S. D., Nicolson G. L. (1994) J. Biol. Chem. 269, 22958–22963 [PubMed] [Google Scholar]
- 17. Dhasarathy A., Wade P. A. (2008) Mutat. Res. 647, 39–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Williams C. J., Naito T., Arco P. G., Seavitt J. R., Cashman S. M., De Souza B., Qi X., Keables P., Von Andrian U. H., Georgopoulos K. (2004) Immunity 20, 719–733 [DOI] [PubMed] [Google Scholar]
- 19. Yoshida T., Hazan I., Zhang J., Ng S. Y., Naito T., Snippert H. J., Heller E. J., Qi X., Lawton L. N., Williams C. J., Georgopoulos K. (2008) Genes Dev. 22, 1174–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pegoraro G., Kubben N., Wickert U., Göhler H., Hoffmann K., Misteli T. (2009) Nat. Cell Biol. 11, 1261–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ramírez J., Hagman J. (2009) Epigenetics 4, 532–536 [DOI] [PubMed] [Google Scholar]
- 22. Miccio A., Wang Y., Hong W., Gregory G. D., Wang H., Yu X., Choi J. K., Shelat S., Tong W., Poncz M., Blobel G. A. (2010) EMBO J. 29, 442–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gao H., Lukin K., Ramírez J., Fields S., Lopez D., Hagman J. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 11258–11263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Grass J. A., Boyer M. E., Pal S., Wu J., Weiss M. J., Bresnick E. H. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 8811–8816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fox A. H., Liew C., Holmes M., Kowalski K., Mackay J., Crossley M. (1999) EMBO J. 18, 2812–2822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Welch J. J., Watts J. A., Vakoc C. R., Yao Y., Wang H., Hardison R. C., Blobel G. A., Chodosh L. A., Weiss M. J. (2004) Blood 104, 3136–3147 [DOI] [PubMed] [Google Scholar]
- 27. Hong W., Nakazawa M., Chen Y. Y., Kori R., Vakoc C. R., Rakowski C., Blobel G. A. (2005) EMBO J. 24, 2367–2378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Leslie A. G. W. (1992) Joint CCP4 + ESF-EAMCB Newsletter on Protein Crystallography, No. 26 [Google Scholar]
- 29. Evans P. (2005) Acta Crystallogr. Sect. D 62, 72–82 [DOI] [PubMed] [Google Scholar]
- 30. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Emsley P., Cowtan K. (2004) Acta. Crystallogr. Sect. D 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 32. Murshudov G., Vagin A., Dodson E. (1997) Acta Crystallogr. Sect. D 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 33. Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B., 3rd, Snoeyink J., Richardson J. S., Richardson D. C. (2007) Nucleic Acids Res. 35, W375–W383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Krissinel E., Henrick K. (2007) J. Mol. Biol. 372, 774–797 [DOI] [PubMed] [Google Scholar]
- 35. DeLano W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientific, Palo Alto, CA [Google Scholar]
- 36. D'Arcy A., Villard F., Marsh M. (2007) Acta Crystallogr. Sect. D 63, 550–554 [DOI] [PubMed] [Google Scholar]
- 37. Song J. J., Garlick J. D., Kingston R. E. (2008) Genes Dev. 22, 1313–1318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lauberth S. M., Rauchman M. (2006) J. Biol. Chem. 281, 23922–23931 [DOI] [PubMed] [Google Scholar]
- 39. Roche A. E., Bassett B. J., Samant S. A., Hong W., Blobel G. A., Svensson E. C. (2008) J. Mol. Cell. Cardiol. 44, 352–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Couture J. F., Collazo E., Trievel R. C. (2006) Nat. Struct. Mol. Biol. 13, 698–703 [DOI] [PubMed] [Google Scholar]
- 41. Han Z., Guo L., Wang H., Shen Y., Deng X. W., Chai J. (2006) Mol. Cell 22, 137–144 [DOI] [PubMed] [Google Scholar]
- 42. Patel A., Vought V. E., Dharmarajan V., Cosgrove M. S. (2008) J. Biol. Chem. 283, 32162–32175 [DOI] [PubMed] [Google Scholar]
- 43. Ruthenburg A. J., Wang W., Graybosch D. M., Li H., Allis C. D., Patel D. J., Verdine G. L. (2006) Nat. Struct. Mol. Biol. 13, 704–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Martin D. I., Orkin S. H.(1990) Genes Dev. 4, 1886–1898 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.