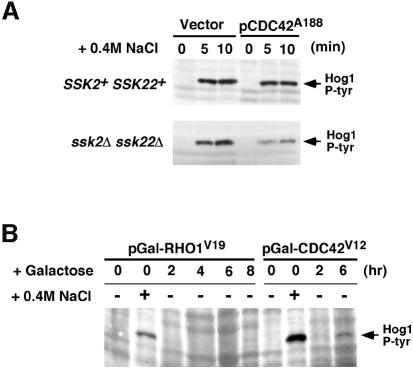
Fig. 9. Effect of dominant-active and dominant-negative CDC42 on Hog1 activation. (A) The dominant-negative CDC42A188 allele inhibits the SHO1 branch of the HOG pathway. Wild-type strain TM141 (SSK2+ SSK22+) and a mutant strain TM252 (ssk2Δ ssk22Δ) were transformed with an expression plasmid pYES2 with a galactose-inducible promoter (Vector), or pYES2 containing the dominant-negative CDC42A188 allele (pCDC42A188). Expression of the CDC42A188 allele was induced by the addition of 2% galactose to exponentially growing cultures containing non-repressing raffinose as carbon source. Two-and-a-half hours after the addition of galactose, the ability of the cells to activate the HOG pathway was determined by monitoring Hog1 tyrosine phosphorylation following osmotic shock. Samples were taken before (time 0), and at 5 and 10 min after the addition of 0.4 M NaCl for 4G10 immunoblot analysis. Expression of the dominant-negative CDC42A188 allele significantly inhibited signaling through the SHO1 branch of the HOG pathway in ssk2Δ ssk22Δ cells. In contrast, activation of Hog1 was unaffected in wild-type cells (SSK2+ SSK22+), in which the SLN1 branch is intact. (B) Expression of the dominant-active CDC42V12 allele activates the HOG pathway in the absence of osmotic stress. Strain TM252 (ssk2Δ ssk22Δ) was transformed with galactose-inducible constructs of dominant-active CDC42V12 or dominant-active RHO1V19. Hog1 activation was assessed as above by 4G10 immunoblot analysis. Exponentially growing cells in non-inducing raffinose media (time 0) were either untreated (–) or treated with NaCl for 5 min (+) as controls for baseline and maximal Hog1 activation, respectively. At the indicated times after addition of 2% galactose to induce expression of RHO1V19 or CDC42V12, samples were harvested (without osmotic shock) for 4G10 analysis. At 6 h of CDC42V12 expression, weak, but significant, phosphorylation of the Hog1 kinase was detected in the absence of osmotic stress.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.