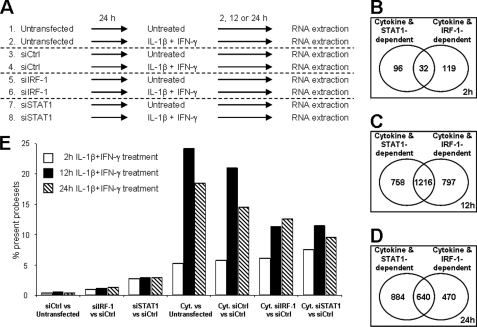
FIGURE 2.
Analysis of gene networks regulated by cytokines, IRF-1, and STAT1 in INS-1E cells. INS-1E cells were left untransfected or transfected with 30 nm of si-control, siIRF-1, or siSTAT1 as described in the legend to Fig. 1. After 24 h of recovery, cells were left untreated or exposed to 10 units/ml IL-1β and 100 units/ml IFN-γ for 2, 12, or 24 h as indicated, before being harvested for RNA extraction and array analysis. A, schematic representation of the microarray conditions (three independent experiments). B–D, Venn diagrams showing the number of β-cell genes whose expression was modified by cytokines and that were either STAT1- or IRF-1-dependent after 2 h (B), 12 h (C), or 24 h (D) of cytokine (Cyt.) exposure. E, mean percentage of probe sets considered as present in the three microarray experiments that were differentially regulated by STAT1/IRF-1 silencing and/or cytokine treatment at 2 h (white bars), 12 h (black bars), or 24 h (hatched bars) of cytokine treatment in the different conditions indicated. Results of three independent array experiments were analyzed. mRNA expression was considered as modified by cytokines when p < 0.02 and fold change >1.5 as compared with untransfected cells not treated with cytokines (Untransfected) or si-control-transfected cells not treated with cytokines (siCtrl).