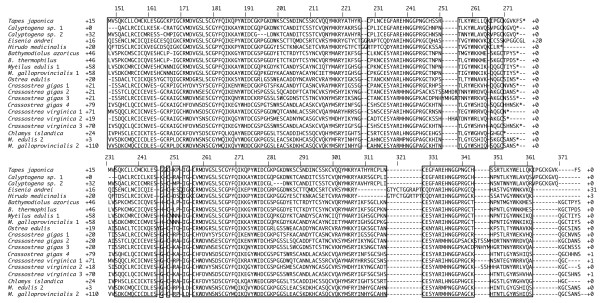
Figure 5.
Amino acid sequence alignment of the i-type lysozymes from bivalve molluscs and annelids. Shown are the alignments of 20 known bivalve i-lysozymes, along with those of two annelids (Eisenia andrei and Hirudo medicinalis) based on ClustalW (top) and BAli-Phy (bottom) algorithms. Boxed sequences indicate the core part of the aligned proteins used for phylogenetic analysis. Residues in front of the position of the first intron are excluded, with the number of residues deleted indicated. The number of residues omitted from the C-terminus is also indicated. The catalytic residues occur at E163 and D174 in the ClustalW (top) alignment. Positional numbering is based on the full alignments.