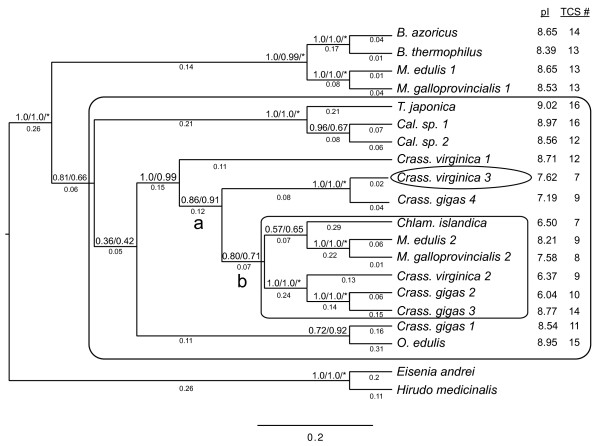
Figure 6.
Phylogenetic relationships among bivalve i-type lysozymes. The phylogenetic tree was inferred based on a Bayesian analysis of amino acid sequences from the core alignment. Numbers above branches indicate Bayesian posterior probabilities, the first for the analysis based on the BAli-Phy alignment and the second for the analysis based on the ClustalW analysis. Asterisks indicate nodes supported by maximum likelihood bootstrap values ≥ 0.75. Branch lengths are shown below branches. The novel C. virginica lysozyme isolated here (cv-lysozyme 3) is circled. The larger box indicates the 13 sequences used for the PAML analysis. The smaller box indicates the clade containing the digestive cv-lysozyme 2 (cv2C). Isoelectric points (pI) and trypsin cutting site numbers (TCS#) predicted from the aligned core sequences are shown.