Abstract
Identification and characterization of the Mycobacterium tuberculosis strains are important for clinical and therapeutic management of tuberculosis. Real-time PCR with a high-resolution melt assay was found to improve the diagnostic process. The assay includes differentiation between M. tuberculosis and Mycobacterium bovis based on one single-nucleotide polymorphism (SNP) in the narGHJI and oxyR genes and determination of M. bovis based on the region of differences 1 (RD1). This assay correctly identified the 7 tested Mycobacterium reference strains and 52 clinical samples with a sensitivity of 2 pg DNA. This assay will help in prescribing adequate treatment and monitoring disease dynamics.
The Mycobacterium tuberculosis complex (MTC) includes Mycobacterium tuberculosis and Mycobacterium africanum, both of which are considered human pathogens, and Mycobacterium microti and Mycobacterium bovis, which are usually associated with animal infections. Although M. tuberculosis is the main cause of human tuberculosis (TB), it has been estimated that M. bovis is responsible for 10 to 15% of new human TB cases in the developing countries (7). The attenuated tuberculosis vaccine strain M. bovis bacillus Calmette-Guérin (BCG) can also cause human TB, especially in patients diagnosed with cellular immunodeficiency (15) or among neonates and children in regions of endemicity who have been vaccinated (5, 7). In 2003, a Palestinian study described an outbreak of BCG complications in the Gaza strip of the Palestinian territories. It affected 225 infants (average age, 4 months), with a complication rate of 36.61 per 1,000 vaccinations (2). Thus, differentiation and identification of M. tuberculosis from other members of the MTC should improve the clinical and therapeutic management of TB. Moreover, differentiation of Mycobacterium species contributes to the understanding of TB epidemiology. Several molecular methods have been reported for genotyping these two pathogens, i.e., multiplex PCR, PCR restriction analysis, allele-specific PCR, and real-time PCR using fluorescent resonance energy transfer (FRET) probes (3, 8, 10, 11, 13). However, these methods are time-consuming, expensive, and complicated. Comparative genome analysis has shown that M. bovis has numerous single-nucleotide polymorphisms (SNPs) in comparison to M. tuberculosis (4). Of these SNPs, the C-to-T transition at position −215 upstream of the GTG start codon in the promoter region of the narGHJI genes was hypothesized to be responsible for the differing nitrate reductase activities between M. tuberculosis and M. bovis (14). Another polymorphic nucleotide was identified in the oxyR gene (11, 12) and was found to be specific for M. bovis. In this study, we describe the use of high-resolution melt curve analysis (HRM) to differentiate between M. bovis and M. tuberculosis by two stepwise reactions. The first reaction is based on the T-to-C transition at position −215 in the promoter region of the narGHJI genes which differentiates between M. tuberculosis and all other members of the MTC (M. africanum, M. microti, and M. bovis) (11). The second reaction is based on the A-to-G polymorphism in the oxyR gene which is specific to M. bovis and which therefore differentiates between M. bovis and M. africanum or M. microti. Such definitive differentiation is essential, even if the organisms are confined to different restricted geographical areas and to different host species. In addition, all M. bovis strains were further identified by targeting the region of differences 1 (RD1; a 9,650-bp deletion which appears as a specific marker for M. bovis BCG) using the HRM assay. The present study was aimed at developing a rapid assay for differentiation between M. bovis and M. tuberculosis with minimal requirements of cost and time.
(This study was submitted by S. Ereqat in partial fulfillment of the requirements for a Ph.D. at Hebrew University under the supervision of Gila Kahila Bar-Gal.)
The study included 52 samples: 7 were previously identified as M. tuberculosis (6) and kindly provided by the Austrian Agency for Health and Food Safety, Vienna, Austria; 15 clinical isolates were recovered from sputum samples at the central laboratories of the Palestinian Ministry of Health; and 30 DNA samples were extracted from Ziehl-Neelsen-stained sputum smears (during the years 2005 to 2009). All archival samples and isolates had been previously identified by IS6110-based PCR to the complex level. Purified DNA from the reference strains M. tuberculosis (H37Rv), M. bovis, and M. bovis BCG and from nontuberculosis mycobacteria (NTM) (M. phlei, M. avium, M. intracellulare, and M. kansasii) were generously provided by the Hebrew University-Hadassah Medical School, Jerusalem, Israel.
Ten samples from TB-negative sputum smears (confirmed negative by IS6110- based PCR) were included as negative controls in the study.
The Ziehl-Neelsen-stained material was scraped off the microscopic slides after the addition of 200 μl of tissue lysis buffer and then processed by proteinase K digestion, followed by extraction according to manufacturer's instructions (High-Pure PCR template preparation; High-Pure, Mannheim, Germany).
A two-reaction approach for differentiating between M. bovis and M. tuberculosis was carried out using two real-time PCR assays followed by HRM analysis. Assay 1 consisted of the identification of the Mycobacterium species based on one SNP, a T-to-C transition, within the narGHJI promoter. Assay 2 consisted of the identification of the Mycobacterium species based on one SNP, an A-to-G transition, of the oxyR gene. The primers used in the two assays were designed to amplify short fragments covering these transitions in both targeted genes. Primer selection was facilitated by Primer3 (http://frodo.wi.mit.edu/). A homology search in GenBank revealed 100% specificity of the primers used for the MTC strains. The primer sequences are shown in Table 1.
TABLE 1.
Primers used in this study
| PCR | Reaction | Gene name | Primer | Primer sequence | Product size (bp) | Reference | Product (Tm [°C])a |
|---|---|---|---|---|---|---|---|
| Preliminary | 1 | narGHJI | LC66 | 5′AACCGACGGTGTGGTTGAC′3 | 155 | Stermann et al. (13) | |
| LC67 | 5′ATCTCGATGGATGGGCGTC′3 | ||||||
| 2 | oxyR | LC90 | 5′CGGGTGCCGCTGACCGCG′3 | 200 | Stermann et al. (13) | ||
| LC91 | 5′CCAGCCGGCTTCGCGTGG′3 | ||||||
| Real-time | 1 | narGHJI | narF | 5′CGCCGTCAACTTGGTTAGA′3 | M. tuberculosis (84.26 ± 0.09) | ||
| narR | 5′GTCCTGCCCGGAAGTTGT′3 | 108 | M. bovis (85.04 ± 0.02) | ||||
| 2 | oxyR | oxyF | 5′ACACTGATTCCGCAGACC′3 | M. bovis (91.9 ± 0.03) | |||
| oxyR | 5′AAAGTCAGCTCTGACAGCGC′3 | 151 | M. bovis (91.27 ± 0.04) |
The product melting temperatures (Tm) are given as means ± standard errors.
Each reaction mixture contained 10 μl of 2× Thermo-Start PCR master mix (Thermo Scientific) and 1.5 μM SYTO 9 (Invitrogen); primer mixes were used in final concentrations of 250 μM. DNA from the clinical isolates and controls was added in 2-μl volumes in total reaction mixtures of 20 μl. The amplification reaction for both amplicons (108 and 151 bp) was done as follows: the temperature was held at 95°C for 15 min for the hot start reaction and then 40 cycles of 5 s of denaturation, 10 s of annealing, and an extension at 55°C with a temperature increase of 0.2°C done were for each step. The melt domain was between 75 and 95°C. Reactions were carried out using the Rotor-Gene 6000 real-time thermal analyzer (Corbett Life Science).
To obtain sufficient DNA for HRM analysis using 14 DNA extracts from Ziehl-Neelsen-stained sputum smears with a low bacterial load, fragments of 200 and 155 bp from the targeted genes were amplified using the previously published primers (13) (Table 1) and then 0.5-μl volumes from the PCR products were subjected to the real-time PCR HRM analysis as described above. In all amplification reactions, we used the negative-control samples to rule out the possibility of PCR contamination. Moreover, each PCR assay included DNA from the reference strains M. tuberculosis (H37Rv), M. bovis, and M. bovis BCG as positive controls to validate the characterization of the tested samples. In addition to the reference strains, seven clinical isolates identified as M. tuberculosis were added to the PCR assays for confirmation. All reactions were performed in duplicate.
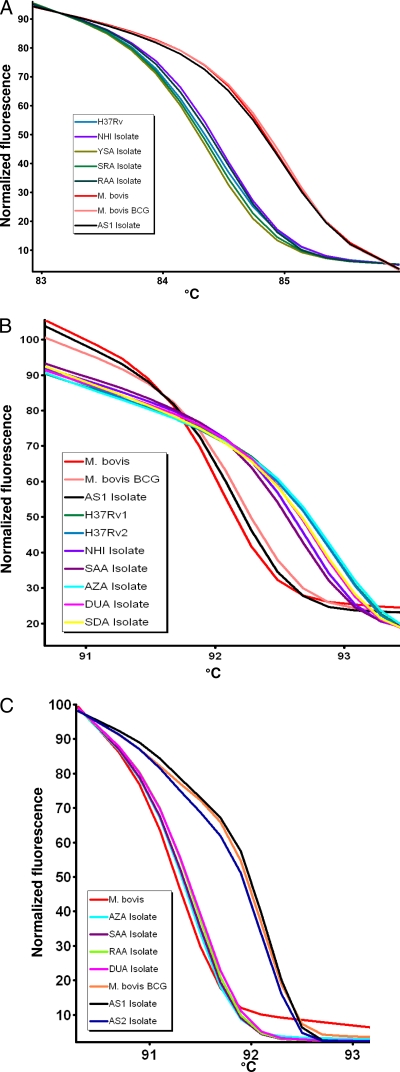
The first step was to optimize the assay, which was carried out on the seven reference samples. The results of the real-time PCR HRM assay based on the narGHJI and oxyR SNPs distinguished between M. tuberculosis isolates and M. bovis, as shown in the normalized melt curves (Fig. 1A and B). The melting temperatures of the PCR product for M. tuberculosis were significantly different from those for M. bovis (P < 0.05) (Table 1). Confirmation of the amplicon size was carried out by agarose gel electrophoresis (data not shown).
FIG. 1.
Characterization of M. tuberculosis and M. bovis based on real-time PCR with high-resolution melt curve analysis identified M. tuberculosis based on the T-to-C transition in the narGHJI genes (A), M. bovis based on the A-to-G transition in the oxyR gene (B), and M. bovis BCG based on the RD1 region (C). For each HRM graph, the x axis shows the temperature in degrees (°C) and the y axis represents the fluorescence signals.
The analytical sensitivity of the HRM assay was clarified by performing 10-fold dilutions using 2 ng of purified DNA for both strains (M. bovis and M. tuberculosis). The melting curve was shown as the DNA level reached 2 pg/reaction. The two reactions showed similar sensitivities. HRM specificity was demonstrated by the absence of melting curves with NTM DNA samples in both reactions.
The second step was to use the optimized assay on the 15 clinical isolates. Fourteen isolates were identified as M. tuberculosis and one isolate as M. bovis using the narGHJI and oxyR SNP assays. The M. bovis isolate was further identified as M. bovis BCG by targeting the RD1 region (13) and applying the HRM assay (Fig. 1C). This isolate was obtained from a 2-year-old female child. The results for the 30 DNA samples extracted from Ziehl-Neelsen-stained sputum smears with unknown Mycobacterium strain identification indicated that 29 samples were M. tuberculosis (their normalized melting curve were identical to those of control strains) and one sample (3%) was M. bovis, which was also obtained from a 3-year-old female child. It is important to note that among the M. tuberculosis strains identified, one was obtained from a nonlocal resident originating from Indonesia, where TB is endemic (imported case), and who was undergoing treatment at the time of sampling (smear positive and culture negative).
The real-time PCR HRM results were confirmed by direct sequencing. The two M. bovis sequences obtained in this study were found to be identical to each other and to the published sequences of M. bovis and M. bovis BCG (GenBank accession no. BX248342.1 and AP010918.1). The sequences of all samples positive for M. tuberculosis (isolates and reference strains) were also found to be 100% identical to each other and to the published M. tuberculosis sequence (GenBank accession no. BX842579.1). Representative DNA sequences (oxyR gene and narGHJI promoter) from patients were deposited in GenBank. The multiple alignment of these DNA sequences is shown in (Fig. 2A and B).
FIG. 2.
(A) Multiple alignment of DNA sequences for the oxyR gene. 1 to 3, sequences belonging to M. bovis (Mb); 4 to 6, sequences belonging to M. tuberculosis (Mt). 1, M. bovis; 2, M. bovis BCG; 3, CaseTBA isolate; 4, CaseASA isolate (accession no. HM135443); 5, M. tuberculosis (H37Rv); 6, CaseSAA isolate (accession no. HM135444); 7, CaseNHI isolate (accession no. HM135445). (B) Multiple alignment of DNA sequences for the narGHJI genes. 1 and 2, sequences belonging to M. tuberculosis (Mt); 3 and 4, sequences belonging to M. bovis (Mb). 1, M. tuberculosis H37Rv; 2: CaseSAA isolate; 3, CaseASA isolate (accession no. HM135442); 4, M. bovis BCG.
The identification of M. bovis among the clinical cases studied is very important, especially for health management. Given that all M. bovis strains are naturally resistant to pyrazinamide (PZA), a first-line antituberculosis agent (1, 7, 9), and that all patients newly diagnosed with TB are placed on a four-drug regimen that includes PZA, definitive differentiation between M. tuberculosis and M. bovis allows for appropriate treatment and reduces the emergence of drug-resistant strains.
The results of this study support the use of a real-time PCR HRM assay on DNA extracted directly from positive Ziehl-Neelsen-stained sputum smear slides. The enrichment of the DNA by PCR prior to the real-time PCR HRM assay was found to be crucial for samples with a low bacterial load. Establishment of this method is of great interest, especially in patients with negative cultures or when the patient is on antitubercular treatment. Determination of the exact strain of Mycobacterium prior to confirmation by culture, which can take up to 3 weeks or more, will improve the treatment by applying the most appropriate drugs. In addition, the possibility of cross-reactivity of the designed primers with other NTM strains was excluded, as none of the tested NTM strains showed any melting curve. The real-time PCR HRM assay developed in this study is less complicated than the assay developed by Stermann and colleagues (13) as it does not require flourescence-labeled probes or combined primers and probes. The simplicity of the assay and its low cost make it applicable for diagnostic laboratories. An advantage of performing HRM analysis on a real-time PCR machine with HRM capability is that the PCR amplification and HRM analysis are performed in the same run and the results are available for assessment of amplification of all samples earlier than if HRM analysis done solely as a quality control measure. Therefore, we recommend this method for the rapid differentiation of M. tuberculosis from M. bovis. This differentiation will aid in choosing an efficient and appropriate treatment for TB patients, will reduce the transmission of the disease, and may prevent further outbreaks.
Nucleotide sequence accession numbers.
Sequences identified in the present study have been deposited in GenBank under accession numbers HM135442, HM135443, HM135444, and HM135445.
Acknowledgments
We thank Asad Ramlawi and Sawsan Said (Palestinian Ministry of Health, Ramallah, Palestinian Authority) for providing the facilities necessary to carry out this work and Herve Bercovier (the Hebrew University-Hadassah Medical School, Jerusalem, Israel) and Werner Ruppitsch (Austrian Agency for Health and Food Safety, Vienna, Austria) for generously providing the Mycobacterium strains.
This study was supported by the German National Science Foundation (Deutsche Forschungsgemeinschaft (DFG grant number NE575/4-1).
Footnotes
Published ahead of print on 15 September 2010.
REFERENCES
- 1.Bakshi, C. S., D. H. Sha, R. Verma, R. K. Singh, and M. Malik. 2005. Rapid differentiation of Mycobacterium bovis and Mycobacterium tuberculosis based on a12.5-Kb fragment by a single tube multiplex PCR. Vet. Microbiol. 109:211-216. [DOI] [PubMed] [Google Scholar]
- 2.Daoud, W. 2003. Control of an outbreak of BCG complications in Gaza. Respirology 8:376-378. [DOI] [PubMed] [Google Scholar]
- 3.Espinosa de los Monteros, L. E., J. Galan, M. Gutierrez, S. Samper, J. F. G. Marin, C. Martin, L. Dominguez, L. de Rafael, F. Baquero, E. G. Mampaso, and J. Blazquez. 1998. Allele-specific PCR method based on pncA and oxyR sequences for distinguishing Mycobacterium bovis from Mycobacterium tuberculosis: intraspecific M. bovis pncA sequence polymorphism. J. Clin. Microbiol. 36:239-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garnier, T., K. Eiglmeier, J. C. Camus, N. Medina, H. Mansoor, M. Pryor, S. Duthoy, S. Grondin, C. Lacroix, C. Monsempe, S. Simon, B. Harris, R. Atkin, J. Doggett, R. Mates, L. Keating, P. R. Wheeler, J. Parkhill, B. G. Barrell, S. T. Cole, S. V. Gordon, and R. G. Hewinson. 2003. The complete genome sequence of Mycobacterium bovis. Proc. Natl. Acad. Sci. U. S. A. 100:7877-7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jou, R., W. L. Huang, and W. J. Su. 2009. Tokyo-172 BCG vaccination complications, Taiwan. Emerg. Infect. Dis. 15:1525-1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pietzka, A. T., A. Indra, A. Stoger, J. Zeinzinger, M. Konard, P. Hasenberger, F. Allerberger, and W. Ruppitsch. 2009. Rapid identification of multidrug-resistant Mycobacterium tuberculosis isolates by rpoB gene scanning using high-resolution melting curve PCR analysis. J. Antimicrob. Chemother. 63:1121-1127. [DOI] [PubMed] [Google Scholar]
- 7.Pinsky, B., and N. Banaei. 2008. Multiplex real-time PCR assay for rapid identification of Mycobacterium tuberculosis complex members to the species level. J. Clin. Microbiol. 46:2241-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rodriguez, J. G., G. J. Mejia, P. D. Portillo, M. E. Patarroyo, and L. A. Murillo. 1995. Species-specific identification of M. bovis by PCR. Microbiology 141:231-2138. [DOI] [PubMed] [Google Scholar]
- 9.Scorpio, A., D. Collin, D. Whipple, D. Cave, J. Bates, and Y. Zhang. 1997. Rapid differentiation of bovine and human tubercle bacilli based on a characteristic mutation in the bovine pyrazinamidase gene. J. Clin. Microbiol. 35:106-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shah, D. H., R. Verma, C. S. Bakshi, and R. K. Singh. 2002. A multiplex-PCR for the differentiation of Mycobacterium bovis and Mycobacterium tuberculosis. FEMS Microbiol. Lett. 214:39-43. [DOI] [PubMed] [Google Scholar]
- 11.Sreevatsan, S., P. Escalante, X. Pan, D. A. Gillies, S. Siddiqi, C. N. Khalaf, B. N. Kreiswirth, P. Bifani, L. G. Adams, T. Fitch, V. S. Perumaalla, M. D. Cave, J. D. van Embden, and J. M. Musser. 1996. Identification of a polymorphic nucleotide in oxyR specific for Mycobacterium bovis. J. Clin. Microbiol. 34:2007-2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sreevatsan, S., S. Pan, Y. Zhang, V. Deretic, and M. Musser. 1997. Analysis of the oxyR-ahpC region in isoniazid-resistant and -susceptible Mycobacterium tuberculosis complex organisms recovered from diseased humans and animals in diverse localities. Antimicrob. Agents Chemother. 41:600-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stermann, M., A. Bohrssen, C. Diephaus, S. Maass, and F. C. Bange. 2003. Polymorphic nucleotide within the promoter of nitrate reductase (NarGHJI) is specific for Mycobacterium tuberculosis. J. Clin. Microbiol. 41:3252-3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stermann, M., L. Sedlacek, S. Maass, and F. Bange. 2004. A promoter mutation causes differential nitrate reductase activity of Mycobacterium tuberculosis and Mycobacterium bovis. J. Bacteriol. 186:2856-2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Talbot, E. A., D. L. Williams, and R. Frothingham. 1997. PCR identification of Mycobacterium bovis BCG. J. Clin. Microbiol. 35:566-569. [DOI] [PMC free article] [PubMed] [Google Scholar]