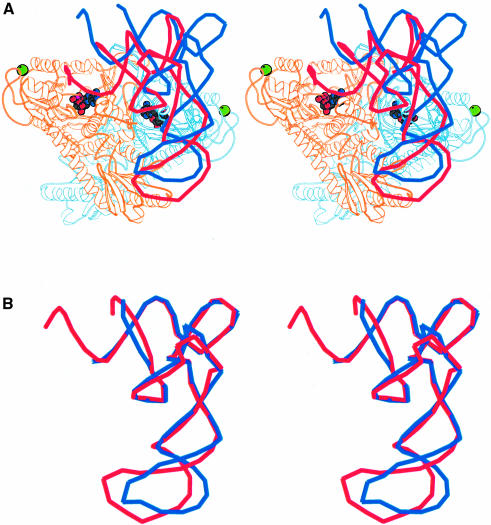
Fig. 6. (A) Stereo diagram of tRNAThr superimposed on the ProRSTT–tRNAPro(CGG)–ProAMS ternary complex. The tRNAThr is coloured red and the tRNAPro is coloured blue. The position of the tRNAThr is that obtained after superimposition of the catalytic domains of ProRSTT and E.coli ThrRS (Sankaranarayanan et al., 1999; PDB entry 1QF6) in their respective tRNA complexes. The red tRNA molecule is thus a good model of tRNAPro functionally bound in the active site of ProRSTT although it does not simultaneously correctly engage with the anticodon-binding domain as the latter is differently orientated with respect to the catalytic domain in E.coli ThrRS. The figure shows that a considerable change in orientation of the tRNAPro from the observed position is required to reach the active conformation (see text). It also suggests that the C-terminal zinc domain could contact the 3′ strand of the acceptor stem. (B) Superimposition of the backbone structures of tRNAPro (blue) and tRNAThr (red) in their respective inactive and active complexes. This shows that, apart from the different distortion of the anticodon loop, the two tRNAs have a remarkably similar conformation. The difference in the anticodon loop is largely due to the fact that base 38 is flipped out in tRNAThr but is base-paired to base 32 in tRNAPro (Sankaranarayanan et al., 1999).

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.