Abstract
Sequence analysis of the Nocardia essential secretory protein SecA1 gene (secA1) for species identification of 120 American Type Culture Collection (ATCC) and clinical isolates of Nocardia (16 species) was studied in comparison with 5′-end 606-bp 16S rRNA gene sequencing. Species determination by both methods was concordant for all 10 ATCC strains. secA1 gene sequencing provided the same species identification as 16S rRNA gene analysis for 94/110 (85.5%) clinical isolates. However, 40 (42.6%) isolates had sequences with <99.0% similarity to archived secA1 sequences for the species, including 29 Nocardia cyriacigeorgica (96.6 to 98.9% similarity) and 4 Nocardia veterana (91.5 to 98.9% similarity) strains. Discrepant species identification was obtained for 16 (14.5%) clinical isolates, including 13/23 Nocardia nova strains (identified as various Nocardia species by secA1 sequencing) and 1 isolate each of Nocardia abscessus (identified as Nocardia asiatica), Nocardia elegans (Nocardia africana), and Nocardia transvalensis (Nocardia blacklockiae); both secA1 gene sequence analysis and deduced amino acid sequence analysis determined the species to be different from those assigned by 16S rRNA gene sequencing. The secA1 locus showed high sequence diversity (66 sequence or genetic types versus 40 16S rRNA gene sequence types), which was highest for N. nova (14 secA1 sequence types), followed by Nocardia farcinica and N. veterana (n = 7 each); there was only a single sequence type among eight Nocardia paucivorans strains. The secA1 locus has potential for species identification as an adjunct to 16S rRNA gene sequencing but requires additional deduced amino acid sequence analysis. It may be a suitable marker for phylogenetic/subtyping studies.
Nocardia spp. are Gram-positive saprophytic bacteria capable of causing suppurative infections, including pulmonary, cutaneous, central nervous system, and disseminated diseases. To date, approximately 90 species have been described (NCBI taxonomy for Nocardia [http://www.ncbi.nlm.nih.gov/Taxonomy/]; http://www.bacterio.cict.fr/n/nocardia.html), at least 33 of which have been implicated in human disease (2). Identification of clinical isolates to the species level is important to characterize associated disease manifestations and to predict antimicrobial susceptibility and for epidemiological and ecological purposes (2, 17).
Because of the difficulty of identifying Nocardia isolates by standard phenotypically based methods and the inability of such methods to identify novel species (2, 17), various nucleic acid amplification methods targeting conserved Nocardia gene regions have been proposed to provide accurate species determination. Of these, sequence analysis of the 16S rRNA gene has become the gold standard for definitive species identification (2, 5, 6, 8, 19). Certain closely related species, however, may not be distinguished by this method due to insufficient interspecies polymorphisms within the 16S rRNA gene sequences (2, 5, 14). Other practical limitations include potential misidentifications as a result of multiple but different copies of the 16S rRNA gene in species such as Nocardia nova (7, 9) and/or the presence of intraspecies 16S rRNA gene sequence polymorphisms (or “sequence types” [STs]) in N. nova, Nocardia cyriacigeorgica, and other species (14, 21).
As such, the continuing evaluation of alternate gene targets to facilitate species identification is important. Sequence polymorphisms within the Nocardia 65-kDa heat shock protein (hsp65), essential secretory protein SecA1 (secA1), gyrase B (gyrB), and 16S-23S rRNA intergenic spacer (ITS) region genes have been reported to enable species level identification (10, 18, 22-24). In particular, sequence variability within a portion (470 bp) of the secA1 gene locus (in conjunction with analysis of deduced amino acid sequences of the SecA1 protein) has shown promise in recognizing and discriminating between the major Nocardia spp. (10). However, data on the application of secA1 gene sequencing in the clinical microbiology laboratory for the identification of Nocardia isolates are few. In one report, reference (n = 30 species), and clinical Nocardia isolates were correctly identified by secA1 gene sequencing (10); in the only other published study, this approach assisted with identification of a novel Nocardia species from soil (16). Evaluation of larger numbers of clinical isolates is essential for establishing a robust repository of secA1 gene sequences.
Our laboratory, which provides regional microbiology services to a large number of health care institutions, has undertaken routine species identification by partial (5′-end 606-bp) 16S rRNA gene sequencing of Nocardia isolates since 2005. In the course of evaluating this approach to providing species identification, we identified significant intraspecies sequence heterogeneity within certain species, such as N. nova and Nocardia brasiliensis (14), highlighting the need to recognize species-specific sequence-based genetic types, or sequence types. Here, to explore the potential of sequence analysis of the secA1 gene as an adjunct to, or a possible substitute for, 16S rRNA gene sequencing, we performed species identification of 120 Nocardia reference and clinical isolates representing the 16 most clinically relevant species by secA1 gene sequence analysis and compared the results with 5′-end 606-bp 16S rRNA gene sequencing. We also report on the genetic diversity of the Nocardia secA1 gene.
MATERIALS AND METHODS
Nocardia organisms.
A total of 120 Nocardia isolates were studied (see Table S1 in the supplemental material). They comprised 10 American Type Culture Collection (ATCC) (Rockville, MD) strains (nine species) (Table 1) and 110 clinical isolates from the Centre for Infectious Diseases and Microbiology Laboratory Services, Westmead Hospital, Sydney, Australia. Clinical isolates were recovered from separate patients from 1997 to 2007. All isolates were identified by standard phenotypic methods and antimicrobial susceptibility profiles (17). The organisms were cultured in brain heart infusion (BHI) broth (Amyl Media, Dandenong, Australia) for 3 to 15 days at 37°C in air prior to analysis by secA1 and 16S rRNA gene sequencing. Sequence analysis of the 16S rRNA gene has been routinely performed in our laboratory since 2005.
TABLE 1.
ATCC reference strains used in the evaluation of secA1 gene sequencing to identify Nocardia spp.
| Species | Strain identification no. | GenBank accession no. |
|
|---|---|---|---|
| 16S rRNA gene | secA1 gene | ||
| N. abscessus | ATCC BAA-279T | AY544980 | DQ360260 |
| N. asteroides | ATCC 19247T | AY756541 | DQ360267 |
| N. brasiliensis | ATCC 19296T | FJ172108 | DQ360269 |
| N. brevicatena | ATCC 15333T | AY756545 | GU595456a |
| N. farcinica | ATCC 3308 | AY756551 | DQ360274 |
| N. farcinica | ATCC 3318T | AY756551 | DQ360274 |
| N. nova | ATCC 33726T | AY756555 | DQ360279 |
| N. paucivorans | ATCC BAA-278T | FJ172128 | GU595459b |
| N. transvalensis | ATCC 6865T | AY756563 | GU179125c |
| N. veterana | ATCC BAA-509 | AY756566 | GU179127d |
For N. brevicatena ATCC 15333T, the secA1 sequence obtained had a C in nucleotide position 469; the corresponding nucleotide is designated Y in the publication by Conville et al. (10) (GenBank accession number DQ360270); there was no amino acid discrepancy resulting from this nucleotide change.
For N. paucivorans ATCC BAA-278T, the secA1 sequence yielded a G at bp 469. The corresponding nucleotide at this position was C for the same strain in the publication by Conville et al. (10) (GenBank accession number DQ360281); there was no resulting amino acid change.
For N. transvalensis ATCC 6865T, the secA1 gene sequence yielded a C in bp position 469 compared to a Y (C/T) for this isolate in the publication by Conville et al. (10) (GenBank accession number DQ360287), but with no resulting amino acid change.
For N. veterana ATCC BAA-509, the secA1 gene sequence yielded a C in bp position 145 and C in bp position 469. The corresponding nucleotides at these positions from the same strain studied by Conville et al. (10) are T and Y (C/T), respectively (GenBank accession number DQ360288), but with no resultant amino acid change.
DNA extraction.
DNA extraction from pure bacterial cultures was performed as previously described (14, 24).
PCR amplification and sequence analysis of the 16S rRNA gene.
Primer design, the PCR parameters employed to amplify the 5′ 606-bp length of the Nocardia 16S rRNA gene, and sequencing of amplified PCR products following their purification were as previously reported (14, 24). 16S rRNA gene sequences were examined using the Biomanager facility (http://biomanager.info/) and aligned against archived sequences in the GenBank database using the BLASTn program (1). In general, a similarity score of ≥99.0% between the unknown sequence and the reference database sequence(s) was used as the criterion to classify an isolate to the species level (5, 19), while a 97 to 98.9% similarity score identified an isolate as belonging to the genus Nocardia but to a different species (4, 12). If the unknown sequence met the criterion for a species but demonstrated nucleotide heterogeneity with the sequence of the reference strain(s) for that species, the sequence was considered to represent a different 16S rRNA gene sequence type (14).
Molecular analysis of the secA1 gene. (i) Primers and PCR amplification.
For all isolates, a 470-bp region of the secA1 gene (corresponding to bp 444 to 913 inclusive of the secA1 gene sequence of Nocardia farcinica IFM 10152) was amplified using secA1-specific primers with tails containing M13 binding sites, as previously described by Conville et al. (10). The sequences of the primers were as follows (the sequences of the tails are indicated by boldface): 5′GTA AAA CGA CGG CCA GGA CAG YGA GTG GAT GGG YCG SGT GCA CCG3′ and 5′CAG GAA ACA GCT ATG ACG CGG ACG ATG TAG TCC TTG TC3′.
Each PCR mixture contained 5 μl template DNA, 0.25 μl (50 pmol/μl) each of forward primer and reverse primer, 1.25 μl deoxynucleoside triphosphates (dNTPs) (2.5 mM each dNTP; Roche Diagnostics, Mannheim, Germany), 2.5 μl 10× PCR buffer (Qiagen, Doncaster, Victoria, Australia), 0.1 μl HotStar Taq polymerase (5 U/μl), and water to a 25-μl final volume. Amplification was performed in a Mastercycler gradient thermocycler (Eppendorf; Netheler-Hinz GmbH, Germany). The cycling conditions were 95°C for 15 min, followed by 35 cycles of 94°C for 30 s, 60°C for 1 min, and 72°C for 1 min, with a final extension step at 72°C for 10 min.
(ii) Sequencing.
PCR products were purified (PCR Product Presequencing Kit; USB Corporation, Cleveland, OH) and sequenced using the BigDye Terminator version 3.1 cycle-sequencing kit (ABI Prism 3100 genetic analyzer; Applied Biosystems, Foster City, CA), and the primers M13-20 forward (5′GTA AAA CGA CGG CCA G3′) and M13 reverse (5′CAG GAA ACA GCT ATG AC3′) (10) were used as sequence primers to obtain double-strand sequencing results. Each sequence was manually aligned and analyzed to ensure high-quality sequence data.
(iii) Assignment of species.
The secA1 sequences obtained in the present study were aligned and compared with those of (i) 30 unique Nocardia type or reference strains (30 species) as previously published (10) and (ii) reference strains of additional Nocardia species (e.g., Nocardia aobensis [GenBank accession no. EU178744], Nocardia blacklockiae [GenBank accession no. EU099362], Nocardia thailandica [GenBank accession no. EU178752], and Nocardia vermiculata [GenBank accession no. EU178753]) archived in the GenBank database. Species identification based on secA1 gene analysis was performed as described by Conville et al. (10).
After examination of sequence traces of each strain, the sequences were manually aligned and analyzed using the Biomanager facility (http://biomanager.info/). SecA1 amino acid sequences were deduced using the Transeq program and compared with archived GenBank sequences using the BLASTp program. Using both BLASTp and BLASTn, the “best-match” results for type/reference strain sequences were defined to decide the species identification of the study isolate; the BLASTp and BLASTn results were also examined for sequence diversity within a species. Where the unknown secA1 gene sequence met the criterion for an individual species (see above) but demonstrated nucleotide heterogeneity with the sequence of the type/reference strain(s) for that species, the sequence was considered to represent a different secA1 sequence type.
Nucleotide sequence accession numbers.
Novel partial secA1 gene sequences (i.e., those with <100% sequence similarity to the type or reference strain in the GenBank database) obtained in this study have been deposited in the GenBank database under accession numbers GU179082 to GU179133 and GU595456 to GU595460 (see Table S1 in the supplemental material). The 16S rRNA gene sequence accession numbers are FJ172102 to FJ172134 (see Table S1 in the supplemental material).
RESULTS
secA1 gene sequences of reference Nocardia isolates.
Sequence analysis of a 470-bp region of the secA1 gene for each of the type strains of Nocardia studied correctly identified all 10 reference strains. The gene sequences showed sufficient base diversity to allow clear species designation of the nine species evaluated (Table 1). The percentage sequence similarity between Nocardia brevitecana and Nocardia paucivorans, two species highly similar by 16S rRNA gene sequencing (99.0%; 600/606 bp) (2), was 95.3% (448/470 bp). For four isolates, N. brevicatena ATCC 15333T, N. paucivorans BAA-278T, Nocardia transvalensis ATCC 6865T, and Nocardia veterana (ATCC BAA-509), sequencing yielded results with 1- or 2-bp differences at the very 3′ end (bp 469), and at bp 145 in one instance, compared to the sequences of the same isolates described previously (10) (Table 1); however, these base pair discrepancies did not result in changes to the deduced amino acid sequences. The secA1 sequences of the remaining six isolates were identical to those already reported (Table 1) and were thus considered to be the same secA1 sequence type (see Materials and Methods and below). Alignment of the deduced amino acid sequences (156 amino acid residues) of the amplified gene region also showed good separation of all reference strains, with the strains of each species showing a unique amino acid sequence (the two ATCC N. farcinica strains had the same sequence [Table 1]). Species identification by secA1 gene sequencing was concordant with partial 16S rRNA gene sequence analysis for all strains.
secA1 gene sequences of clinical isolates.
The secA1 gene sequence and the deduced amino acid sequence data in comparison to results obtained by partial 16S rRNA gene sequencing of 110 clinical Nocardia isolates belonging to 15 species are shown in Tables 2 and 3.
TABLE 2.
Species identifications of 94 clinical Nocardia isolates by secA1 gene sequencing where species assignment was concordant with partial 16S rRNA gene sequencing
| Species identification (606-bp 16S rRNA gene) | No. of isolates | Sequence similarity to reference sequences |
||
|---|---|---|---|---|
| secA1 gene [bp (%)] | SecA1 amino acid [bp (%)] | 606-bp 16S rRNA gene (%) | ||
| N. abscessus | 1 | 469/470 (99.8) | 156/156 (100) | 100 |
| N. arthritidis | 1 | 463/470 (98.5) | 156/156 (100) | 604/606 (99.7) |
| N. asteroides | 1 | 470/470 (100) | 156/156 (100) | 100 |
| 1 | 458/470 (97.4) | 156/156 (100) | 600/606 (99.0) | |
| N. beijingensis | 1 | 469/470 (99.8) | 156/156 (100) | 100 |
| N. brasiliensis | 2 | 466/470 (99.1) | 155/156 (99.4) | 604/606 (99.7) |
| 2 | 465/470 (98.9) | 155/156 (99.4) | 604/606 (99.7) | |
| 1 | 464/470 (98.7) | 155/156 (99.4) | 604/606 (99.7) | |
| 1 | 459/470 (97.6) | 154/156 (98.7) | 603/606 (99.5) | |
| 1 | 460/470 (97.9) | 154/156 (98.7) | 602/606 (99.3) | |
| N. cyriacigeorgica | 3 | 470/470 (100) | 156/156 (100) | 100 |
| 2a | 467/470 (99.4) | 156/156 (100) | 100 | |
| 1 | 465/470 (98.9) | 156/156 (100) | 100 | |
| 1 | 463/470 (98.5) | 153/156 (98.1) | 100 | |
| 1 | 461/470 (98.1) | 155/156 (99.4) | 100 | |
| 10a | 459/470 (97.6) | 156/156 (100) | 100 | |
| 7a | 458/470 (97.4) | 155/156 (99.4) | 100 | |
| 1 | 457/470 (97.2) | 155/156 (99.4) | 100 | |
| 1 | 456/470 (97.0) | 154/156 (98.7) | 100 | |
| 1 | 454/470 (96.6) | 154/156 (98.7) | 100 | |
| 1 | 459/470 (97.6) | 156/156 (100) | 605/606 (99.8) | |
| 5a | 458/470 (97.4) | 155/156 (98.7) | 605/606 (99.8) | |
| N. farcinica | 7a | 469/470 (99.8) | 156/156 (100) | 100 |
| 5a | 468/470 (99.6) | 156/156 (100) | 100 | |
| 1 | 467/470 (99.4) | 156/156 (100) | 100 | |
| 1 | 468/470 (99.6) | 156/156 (100) | 605/606 (99.8) | |
| 1 | 468/470 (99.6) | 156/156 (100) | 604/606 (99.7) | |
| N. nova | 4 | 470/470 (100) | 156/156 (100) | 100 |
| 2 | 469/470 (99.8) | 156/156 (100) | 100 | |
| 1 | 470/470 (100) | 156/156 (100) | 605/606 (99.8) | |
| 1 | 467/470 (99.4) | 156/156 (100) | 604/606 (99.7) | |
| 1 | 466/470 (99.1) | 156/156 (100) | 604/606 (99.7) | |
| 1 | 470/470 (100) | 156/156 (100) | 602/606 (99.3) | |
| N. otitidiscaviarum | 2 | 470/470 (100) | 156/156 (100) | 100 |
| 1 | 469/470 (99.8) | 156/156 (100) | 100 | |
| 1 | 467/470 (99.4) | 156/156 (100) | 100 | |
| N. paucivorans | 7 | 469/470 (99.8) | 156/156 (100) | 606/608 (99.7) |
| N. thailandica | 1 | 467/468 (99.8) | 155/156 (98.7) | 100 |
| N. veterana | 2 | 469/470 (99.8) | 156/156 (100) | 100 |
| 2 | 468/470 (99.6) | 156/156 (100) | 100 | |
| 2 | 467/470 (99.4) | 156/156 (100) | 100 | |
| 1 | 465/470 (98.9) | 156/156 (100) | 100 | |
| 1 | 437/470 (93.0) | 154/156 (98.7) | 100 | |
| 1b | 430/470 (91.5) | 155/156 (99.4) | 599/606 (98.8) | |
| 1c | 437/470 (93.0) | 155/156 (99.4) | 596/606 (98.3) | |
| N. vinacea | 1 | 466/470 (99.1) | 156/156 (100) | 100 |
Strains with the same percentage sequence similarity but with one or more strains yielding nucleotide heterogeneity and hence representing a different sequence type of the secA1 gene(see Table S1 in the supplemental material).
The 16S rRNA gene sequence was 98.8% similar to archived N. veterana 16S rRNA gene sequences in the GenBank database. SecA1 amino acid analysis identified the isolate as N. veterana.
The 16S rRNA gene sequence was 98.3% similar to archived N. veterana 16S rRNA gene sequences in the GenBank database. SecA1 amino acid analysis identified the isolate as N. veterana.
TABLE 3.
Discordant species identification of 16 clinical Nocardia isolates by secA1 gene sequencing in comparison with 16S rRNA gene sequence analysis
| Species identification |
No. of isolates | GenBank accession no. (secA1 gene)a | No. of bp differences from reference secA1 sequence |
No. of amino acid differences from reference SecA1 sequence |
|||
|---|---|---|---|---|---|---|---|
| 16S rRNA gene analysis | secA1 gene analysis | secA1-based species identificationb | 16S rRNA-based species identificationc | secA1-based species identificationd | 16S rRNA-based species identificatione | ||
| N. abscessus | N. asiatica | 1 | GU179083 | 2 | 31 | 0 | 2 |
| N. elegans | N. africana | 1 | GU179105 | 3 | 8 | 0 | 3 |
| N. nova | N. aobensis | 1 | GU595458 | 2 | 10 | 1 | 3 |
| N. nova | N. aobensis | 1 | GU179112 | 1 | 6 | 0 | 2 |
| N nova | N. aobensis | 2 | GU179114 | 2 | 8 | 0 | 2 |
| N. nova | N. aobensis | 1 | GU179117 | 4 | 8 | 0 | 2 |
| N. nova | N. brasiliensis | 1 | DQ360269 (=) | 0 | 52 | 0 | 7 |
| N. nova | N. elegans | 3 | GU179115 | 4 | 4 | 0 | 1 |
| N. nova | N. elegans | 1 | GU179116 | 4 | 5 | 0 | 1 |
| N. nova | N. kruczakiae | 1 | GU179118 | 10 | 12 | 1 | 3 |
| N. nova | N. veterana | 1 | GU179119 | 37 | 39 | 1 | 4 |
| N. nova | N. veterana | 1 | GU179128 (=) | 33 | 37 | 0 | 3 |
| N. transvalensis | N. blacklockiae | 1 | GU179126 | 8 | 18 | 1 | 3 |
GenBank accession numbers of secA1 sequence results generated in the present study. =, 100% sequence identity with an existing GenBank reference sequence for the species.
Number of bp differences between the obtained secA1 gene sequences in the present study and the best-match secA1 sequence of the reference strain(s) for the species as determined by secA1 analysis.
Number of bp differences between the secA1 gene sequences in the present study and the best-match secA1 sequence of the reference strains(s) for the species as designated by 606-bp 16S rRNA sequencing.
Number of deduced amino acid differences between the SecA1 protein sequences obtained in the present study and the best-match reference protein sequence for the species as determined by secA1 analysis.
Number of deduced amino acid differences between the SecA1 protein sequences obtained in the present study and the best-match secA1 sequences of the reference strain(s) for the species as determined by 16S rRNA sequencing.
By 16S rRNA gene sequencing (14, 24), there were 34 isolates of N. cyriacigeorgica, 23 of N. nova, and 15 of N. farcinica; seven strains each of N. paucivorans and N. brasiliensis and four of Nocardia otitidiscavarium; and two strains each of Nocardia asteroides sensu stricto and Nocardia abscessus. The remaining species were Nocardia arthriditis, Nocardia elegans, N. thailandica, and Nocardia vinacea (each n = 1) and 10 strains assigned to N. veterana. Of the 10 N. veterana isolates, 8 had 16S rRNA gene sequences with ≥99.0% similarity to reference N. veterana sequences (14) and 2 (strains 05-144-3166 and 07-296-2401 [see Table S1 in the supplemental material]) had 98.3% and 98.8% sequence similarity, respectively; because these sequences were most similar to N. veterana 16S rRNA reference sequences, these two isolates were determined to be N. veterana (Table 2).
Species identification concordant with 16S rRNA gene sequencing.
Table 2 summarizes the results for 94/110 isolates for which species identification by secA1 and 16S rRNA gene sequencing were concordant (85.5%). secA1 gene sequencing was able to correctly identify most major Nocardia species, with the exception of N. nova (see “Discrepant species identification between 16S rRNA gene and secA1 gene sequencing” below). The sequences of 54 of the 94 (57.4%) isolates showed ≥99% similarity to secA1 gene sequences of the type or reference strain of the species to which they were determined to belong when analyzed by 16S rRNA gene sequencing (Table 2), as previously published (10). Twenty-nine of 34 N. cyriacigeorgica strains, 1 of 2 N. asteroides strains, and 5 of 7 N. brasiliensis isolates, however, yielded secA1 sequences that showed <99% (96.6 to 98.9%) gene sequence similarity to the type/reference strains of their respective species (Table 2), and 3 of 10 N. veterana isolates had sequences with only 91.5 to 93.0% similarity to the reference N. veterana secA1 sequence in the GenBank database (GenBank accession no. DQ360288). For example, the secA1 sequence of N. veterana strain 07-296-2401 (see Table S1 in the supplemental material; GenBank accession no. GU179131) had 91.5% sequence similarity (430/470 bp) to the reference N. veterana strain (GenBank accession no. DQ360288) but yielded 94.0% sequence similarity (442/470 bp) to the N. nova sequence (GenBank accession no. DQ360279). However, because its deduced SecA1 amino acid sequence was most similar to that known for N. veterana (155/156 amino acids; 99.4% match), the isolate was determined to be N. veterana. Similarly, the other two N. veterana strains with 93.0% secA1 gene sequence similarity to the sequence of the reference N. veterana strain were determined to belong to that species based on a deduced amino acid sequence similarity with 1 or 2 amino acid differences (Table 2).
The overall, sequence diversity within species for the region of the secA1 gene studied ranged from 0 to 40 bp (Table 2), while the deduced amino acid sequences of 93 isolates were between 98.7% and 100% similar to that of the type/reference strain (0 to 2 amino acid differences), with only one isolate (an N. cyriacigeorgica strain) yielding a 3-amino-acid difference (Table 2). This is well illustrated by the results obtained for the three N. veterana strains (see above) and for an isolate of N. asteroides (Table 2). Alignment of the deduced amino acid sequences provided accurate species identification for all 94 isolates.
Discrepant species identification between 16S rRNA gene and secA1 gene sequencing.
Discrepant species identification by secA1 and 16S rRNA gene sequencing was observed for 16 isolates (Table 3; see Table S1 in the supplemental material). This was most evident for N. nova; of a total of 23 clinical strains studied, 13 were determined to be other species by secA1 gene sequencing. They were N. aobensis (n = 5 strains), N. elegans (n = 4), N. veterana (n = 2), N. brasiliensis (n = 1), and Nocardia kruczakiae (n = 1). Other discrepant results involved a single strain each of N. abscessus, N. elegans, and N. transvalensis (species determination by 16S rRNA sequence analysis) identified by secA1 sequencing as Nocardia asiatica, Nocardia africana, and N. blacklockiae, respectively (Table 3). In all cases, the isolates had greater sequence similarity to the species determined when the secA1 gene sequence and amino acid sequences were evaluated (0 or 1 amino acid difference; 0 to 37 bp differences) than to the species as determined by 16S rRNA gene sequencing (1 to 7 amino acid differences; 4 to 39 bp differences) (Table 3). For example, the isolate identified as N. elegans by 16S rRNA gene sequencing (100% sequence similarity) yielded secA1 sequences with greater similarity to N. africana (99.4% [467/470 bp]) than to N. elegans (98.3% [462/470 bp]; in addition, the deduced amino acid sequence was identical to the amino acid sequence of the type strain of N. africana but showed a 3-amino-acid difference from the sequence of the type strain of N. elegans).
Intraspecies secA1 gene and SecA1 amino acid sequence polymorphisms.
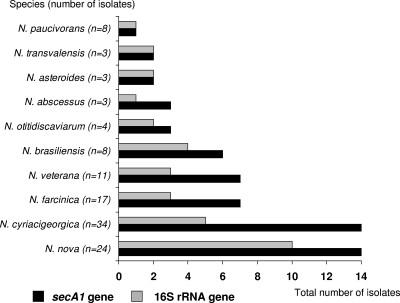
For clinical and type/reference isolates belonging to the same species, secA1 gene sequence diversity was greater than the sequence diversity previously seen with the 16S rRNA gene sequences (Fig. 1) (14). For the nine species represented by >1 isolate, intraspecies diversity was most evident among N. nova (14 secA1 STs or polymorphisms among 24 strains, including that observed for the type strains), followed by N. cyriacigeorgica (14 STs; 34 strains), N. farcinica and N. veterana (n = 7 each), and N. brasiliensis (6 STs among 8 strains) (Fig. 1). In contrast, there was a single ST among eight strains of N. paucivorans. The relative proportions of STs according to species corresponded in general to those of 16S rRNA sequence types. Overall, the number of secA1 STs (n = 66) exceeded that of SecA1 amino acid STs (n = 39) and of 16S rRNA gene types (n = 40) (Fig. 1; see Table S1 in the supplemental material). Furthermore, the combined genetic diversity at the secA1 and 16S rRNA loci enabled the identification of the same or a greater (N. farcinica, N. nova, and N. cyriacigeorgica) number of STs than that using either locus alone. Of note, for N. abscessus, the secA1 locus was the only contributor to the genetic diversity; all three strains had identical 16S rRNA gene sequences but three different secA1 sequences (see Table S1 in the supplemental material).
FIG. 1.
Genetic types or sequence types of Nocardia by secA1 and partial 16S rRNA gene sequence analysis according to species. The number of strains is provided on the y axis and the number of sequence types on the x axis.
DISCUSSION
Species identification of Nocardia organisms remains a challenge despite advances in molecularly based identification methods. The results of the present study show that secA1 gene sequence analysis in combination with deduced amino acid sequence results enabled accurate identification of a large number of clinical isolates (94/110) of the medically important genus Nocardia to species level, including differentiation between closely related species (e.g., N. brevitecana and N. paucivorans; N. nova and N. veterana). Other key findings include the substantial intraspecies genetic diversity within this portion of the Nocardia secA1 gene (with 66 different secA1 STs among 120 isolates) in comparison with the diversity within the 16S rRNA gene. By studying such STs, particularly in combination with 16S rRNA gene polymorphisms, a “barcoding” or dual-locus genotyping approach may potentially be useful, not only for species identification, but as an epidemiological tool.
Validation of sequence-based identification methods using large numbers of isolates with well-characterized phenotypic, as well as genetic, traits is essential to determine their utility in the diagnostic microbiology laboratory. By comparison with partial (5′-end 606-bp) 16S rRNA gene sequencing, secA1 gene analysis provided clear species identification of all 10 ATCC Nocardia strains (sensitivity, 100%). The explanation for the 1- or 2-bp nucleotide differences between the isolates and the same (type) strain tested previously (Table 1) is not known; evaluation of the deduced amino acid sequences of the SecA1 protein supported the same species designation.
Among 110 isolates identified to the species level by partial 16S rRNA gene sequencing, 94 (85.5%) yielded identical species identification by secA1 gene sequencing (Table 2). Although a large proportion (57.4%) of these isolates demonstrated >99.0% similarity to the corresponding type strain with identical amino acid sequences, 33 strains showed 10- to 40-bp differences from their respective type strains, most notably three N. veterana isolates, which produced only 91.5 to 93.0% sequence similarity. The reasons for the sequence discrepancy between the study N. veterana isolates and the N. veterana type strain (10) remain unclear, but our findings are consistent with the substantial genetic diversity within the secA1 gene locus (see below) and highlight the importance of evaluation of larger numbers of isolates of multiple species in creating a robust library of sequences. Nonetheless, analysis of the deduced amino acid sequences of the SecA1 protein gave unambiguous identification in all the cases mentioned above. Collectively, then, and especially where limited numbers of isolates are available for study, evaluation of amino acid sequences is important for accurate species identification, as has been previously suggested (10); however, further study with additional clinical isolates is necessary to verify this conclusion.
In contrast to the findings of Conville et al. (10), a greater proportion (16 of 110 versus 1 of 40) of clinical isolates yielded secA1 gene sequences (both deduced amino acid and DNA sequences) that resulted in discrepant species assignment compared with that determined by partial 16S rRNA sequencing (Table 3). This was most problematic for N. nova, where as many as 52 nucleotide differences between the sequence being queried and the reference strain's sequence were identified. Although secA1 gene and amino acid sequence analyses demonstrated inconsistencies between the identification of strains and those (i.e., species identification) based on 16S rRNA gene sequencing, DNA-DNA hybridization studies may be needed to resolve species identification. All “discrepant” identification results occurred between species known to be very closely related (e.g., N. abscessus and N. asiatica) and that in many instances were/are classified within the same species complex (Table 3) (2). One hypothesis is that the secA1 gene, which is a single-copy protein gene, may be affected by lateral gene transfer, resulting in transfer of secA1 gene material to the parent organism from other Nocardia species (i.e., other than its own species). Further examination of this possibility, in the context not only of secA1 gene diversity but of other single-copy Nocardia genes, such as hsp65 (18) is worthy of consideration.
Gene polymorphisms within alternate molecular targets, including gyrB and the 16S-23S rRNA ITS region (13, 22-24), have also been reported to improve species identification, yet the selection of the gene target as an adjunct to, or even a substitute for, 16S rRNA gene sequencing for the identification of Nocardia spp. requires careful consideration. Single-copy gene targets avoid potential misidentifications caused by multiple different copies of the 16S rRNA gene and ITS but suffer from inherent lower sensitivities (13, 18, 22-24). Expanded databases of these gene sequences from clinical isolates are further required to ensure appropriate evaluation of these loci as suitable targets for species identification. The utility of any approach must be tested with a large number of clinical specimens and in different laboratories.
A major finding of the present study was the substantial genetic diversity within species in the secA1 gene locus, exceeding what was previously suggested by partial 16S rRNA gene sequencing (14). Overall, there were a total of 66 secA1 STs compared with 40 sequence-based genetic types of partial 16S rRNA sequences (Fig. 1; see Table S1 in the supplemental material), although the number of SecA1 amino acid sequence types (n = 39) was similar to the number of types of 16S rRNA gene sequences. Our results are consistent with the suggestions that the evolutionary clock of the secA1 gene is faster than that of the 16S rRNA gene (13) and that the 16S rRNA gene and SecA1 amino acid sequences have similar evolution speeds (13, 15). The degree of intraspecies genetic diversity varied with species and in general was correlated with intraspecies heterogeneity for the 16S rRNA gene locus (14); the numbers of secA1 STs were greatest for N. nova, N. cyriacigeorgica, and N. brasiliensis.
The genetic diversity in the secA1 gene identified in this study between and within species can be useful for defining phylogenetic relationships among species and for epidemiological studies (10, 13). Both the secA1 and 16S rRNA gene sequences appeared to be good candidate markers for these considerations (reference 14 and this study). Others have also determined that there are several 16S rRNA genetic types within N. cyriacigeorgica (21). Comparison of STs of isolates from different geographic regions may provide relevant clinical or epidemiological associations. Because secA1 and 16S rRNA genetic types did not always correlate (see Table S1 in the supplemental material), the combination of genetic types encompassing both loci could potentially generate a useful and discriminatory “barcode” or “multinucleotide dual-locus” typing system as an epidemiological tool to be used in conjunction with relevant phenotypic features (3, 11). This approach has been successfully applied to classify eukaryotic organisms, including fungi (20), but has not yet been extended to similar purposes in bacteria (14).
In conclusion, given the potential limitations of 16S rRNA sequence analysis for species identification of Nocardia, continuing evaluation of alternate gene loci to assist with species determination remains important. The secA1 gene may ultimately be most useful as part of a multigene or polyphasic approach to the identification of medically important Nocardia isolates. Analysis of a larger number of species and isolates representing each species is required to ascertain the utility of secA1 gene polymorphisms for reliable species identification.
Supplementary Material
Acknowledgments
We thank the Westmead Millenium Institute Sequencing Laboratory for assistance with gene sequencing.
Footnotes
Published ahead of print on 1 September 2010.
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown-Elliott, B. A., J. M. Brown, P. S. Conville, and R. J. Wallace, Jr. 2006. Clinical and laboratory features of the Nocardia spp. based on current molecular taxonomy. Clin. Microbiol. Rev. 19:259-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.CBOL Plant Working Group. 2009. A DNA barcode for land plants. Proc. Natl. Acad. Sci. U. S. A. 106:12794-12797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clarridge, J. E., III. 2004. Impact of 16S rRNA gene sequence analysis for identification of bacteria on clinical microbiology and infectious diseases. Clin. Microbiol. Rev. 17:840-862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cloud, J. L., P. S. Conville, A. Croft, D. Harmsen, F. G. Witebsky, and K. C. Carroll. 2004. Evaluation of partial 16S ribosomal DNA sequencing for identification of Nocardia species by using the MicroSeq 500 system with an expanded database. J. Clin. Microbiol. 42:578-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Conville, P. S., S. H. Fischer, C. P. Cartwright, and F. G. Witebsky. 2000. Identification of Nocardia species by restriction endonuclease analysis of an amplified portion of the 16S rRNA gene. J. Clin. Microbiol. 38:158-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Conville, P. S., and F. G. Witebsky. 2007. Analysis of multiple differing copies of the 16S rRNA gene in five clinical isolates and three type strains of Nocardia species and implications for species assignment. J. Clin. Microbiol. 45:1146-1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Conville, P. S., J. M. Brown, A. G. Steigerwalt, J. W. Lee, V. L. Anderson, J. T. Fishbain, S. M. Holland, and F. G. Witebsky. 2004. Nocardia kruczakiae sp. nov., a pathogen in immunocompromised patients and a member of the “N. nova complex.” J. Clin. Microbiol. 42:5139-5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Conville, P. S., and F. G. Witebsky. 2005. Multiple copies of the 16S rRNA gene in Nocardia nova isolates and implications for sequence-based identification procedures. J. Clin. Microbiol. 43:2881-2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Conville, P. S., A. M. Zelazny, and F. G. Witebsky. 2006. Analysis of secA1 gene sequences for identification of Nocardia species. J. Clin. Microbiol. 44:2760-2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fazekas, A. J., K. S. Burgess, P. R. Kesanakurti, S. W. Graham, S. G. Newmaster, B. C. Husband, D. M. Percy, M. Hajibabaei, and S. C. Barrett. 2008. Multiple multilocus DNA barcodes from the plastid genome discriminate plant species equally well. PLoS One 3:e2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Janda, J. M., and S. L. Abbott. 2007. 16S rRNA gene sequencing for bacterial identification in the diagnostic laboratory: pluses, perils, and pitfalls. J. Clin. Microbiol. 45:2761-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kang, Y., K. Takeda, K. Yazawa, and Y. Mikami. 2009. Phylogenetic studies of Gordonia species based on gyrB and secA1 gene analyses. Mycopathologia 167:95-105. [DOI] [PubMed] [Google Scholar]
- 14.Kong, F., S. C. Chen, X. Chen, V. Sintchenko, C. Halliday, L. Cai, Z. Tong, O. C. Lee, and T. C. Sorrell. 2009. Assignment of reference 5′-end 16S rDNA sequences and species-specific sequence polymorphisms improves species identification of Nocardia. Open Microbiol. J. 3:97-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuo, C. H., and H. Ochman. 2009. Inferring clocks when lacking rocks: the variable rates of molecular evolution in bacteria. Biol. Direct. 4:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lamm, A. S., A. Khare, P. Conville, P. C. Lau, H. Bergeron, and J. P. Rosazza. 2009. Nocardia iowensis sp. nov., an organism rich in biocatalytically important enzymes and nitric oxide synthase. Int. J. Syst. Evol. Microbiol. 59:2408-2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McNeil, M. M., and J. M. Brown. 1994. The medically important aerobic actinomycetes: epidemiology and microbiology. Clin. Microbiol. Rev. 7:357-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rodríguez-Nava, V., A. Couble, G. Devulder, J. P. Flandrois, P. Boiron, and F. Laurent. 2006. Use of PCR-restriction enzyme pattern analysis and sequencing database for hsp65 gene-based identification of Nocardia species. J. Clin. Microbiol. 44:536-546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roth, A., S. Andrees, R. M. Kroppenstedt, D. Harmsen, and H. Mauch. 2003. Phylogeny of the genus Nocardia based on reassessed 16S rRNA gene sequences reveals underspeciation and division of strains classified as Nocardia asteroides into three established species and two unnamed taxons. J. Clin. Microbiol. 41:851-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Santamaria, M., S. Vicario, G. Pappada, G. Scioscia, C. Scazzocchio, and C. Saccone. 2009. Towards barcode markers in Fungi: an intron map of Ascomycota mitochondria. BMC Bioinform. 10(Suppl. 6):S15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schlaberg, R., R. C. Huard, and P. Della-Latta. 2008. Nocardia cyriacigeorgica, an emerging pathogen in the United States. J. Clin. Microbiol. 46:265-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takeda, K., Y. Kang, K. Yazawa, T. Gonoi, and Y. Mikami. 2010. Phylogenetic studies of Nocardia species based on gyrB gene analyses. J. Med. Microbiol. 59:165-171. [DOI] [PubMed] [Google Scholar]
- 23.Wang, X., M. Xiao, F. Kong, V. Sintchenko, H. Wang, B. Wang, S. Lian, T. Sorrell, and S. C. Chen. 2010. Reverse line blot hybridization (RLB) and DNA sequencing studies of the 16S-23S rRNA gene intergenic spacer (ITS) regions of five emerging pathogenic Nocardia species. J. Med. Microbiol. 59:548-555. [DOI] [PubMed] [Google Scholar]
- 24.Xiao, M., F. Kong, T. C. Sorrell, Y. Cao, O. C. Lee, Y. Liu, V. Sintchenko, and S. C. Chen. 2010. Identification of pathogenic Nocardia species by reverse line blot hybridization targeting the 16S rRNA and 16S-23S rRNA gene spacer regions. J. Clin. Microbiol. 48:503-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.