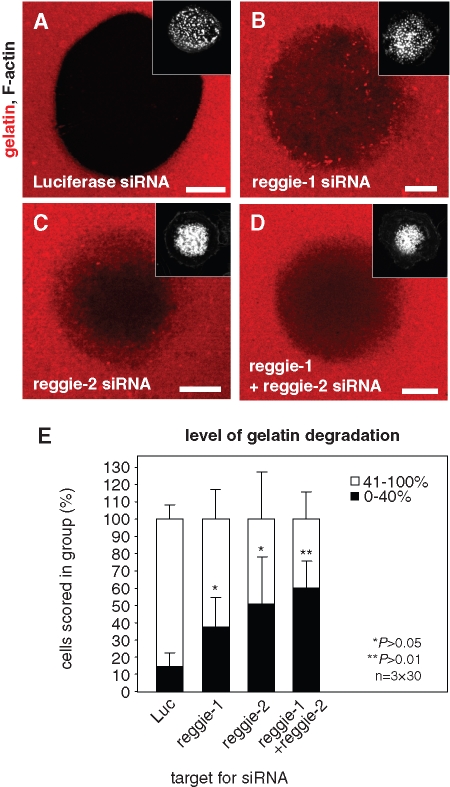
FIGURE 10:
Knockdown of reggie proteins influences matrix degradation. Confocal laser scanning micrographs of primary human macrophages transfected with siRNA-luciferase (A), siRNA-reggie-1 (B), siRNA-reggie-2 (C), or a combination of both reggie siRNAs (D), seeded on rhodamine-labeled gelatin matrix (red). Matrix degradation is visible as dark areas; insets show relevant F-actin staining by Cy5-labeled phalloidin (white). White bar, 10 μm. (E) Evaluation of matrix degradation in cells treated with siRNAs. The degree of matrix degradation was analyzed by fluorescence measurements of 3 × 30 cells each time. Complete absence of labeled matrix beneath cells was set as 100% degradation. Cells were scored into groups according to matrix degradation (0–40% and 41–100%). For differences between control values and values gained with reggie siRNAs, a P value < 0.05 was considered significant, and a P value < 0.01 was considered highly significant (indicated by asterisks). Values are given as mean percentage ± SD of total counts in Table 1.