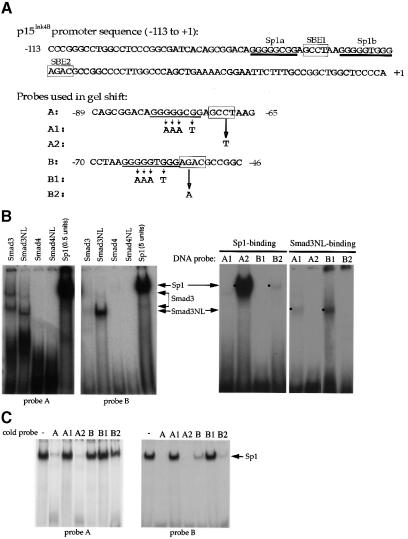
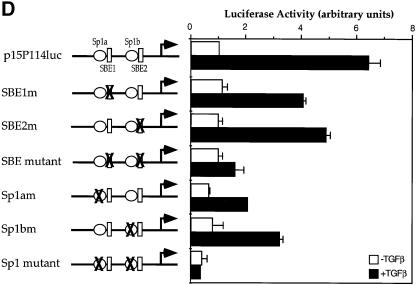
Fig. 3. Sp1 binding sites and Smad3-binding elements are required for the activation of the p15Ink4B gene. (A) Nucleotide sequence of the –113 to +1 segment of the p15Ink4B promoter. Predicted Sp1 binding sites (Sp1a and Sp1b) and SBEs 1 and 2 are indicated. The oligonucleotide probes A and B, used in the gel shift experiments, with their inactivating mutations are also shown. (B) Sp1 and Smad3 bind to the p15Ink4B promoter. Purified Sp1 (0.5 or 5 U) or GST–Smad fusion proteins (1 µg) were incubated with the 32P-labeled probe A or B, or the mutant probes A1 or B1, in which the Sp1a or Sp1b sites are mutated, or the mutant A2 or B2 probes, in which the SBE1 or SBE2 sites are inactivated. Gel-shifted DNA–protein complexes are marked. (C) Sp1 binding to the Sp1 binding sites in oligonucleotides A or B can be competed with unlabeled ‘wild-type’ oligonucleotides, but not by oligonucleotides with mutated Sp1 binding sites. Purified Sp1 was incubated with the 32P-labeled probe A or B in the presence of indicated unlabeled DNA. The DNA–Sp1 complex is marked. (D) Mutational analysis of the p15Ink4B promoter. Sp1 sites and SBEs were individually, or in combination, mutated in the p15Ink4B promoter, as shown in Figure 3A. Transcription was measured by luciferase activity. Transfection, TGF-β treatment and luciferase assay in HaCaT cells were performed as in Figure 1.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.