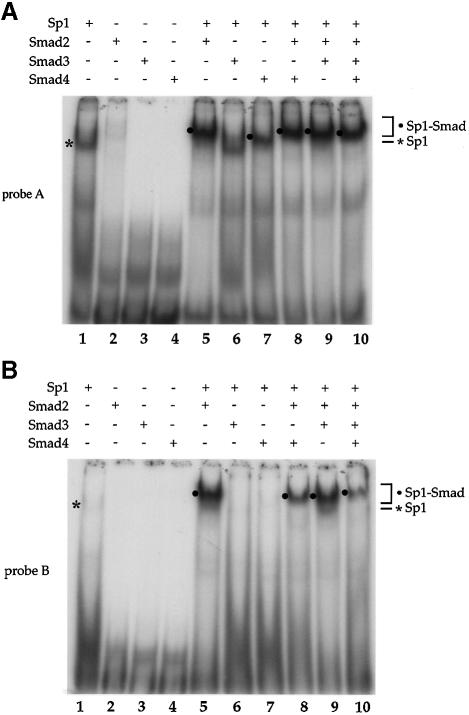
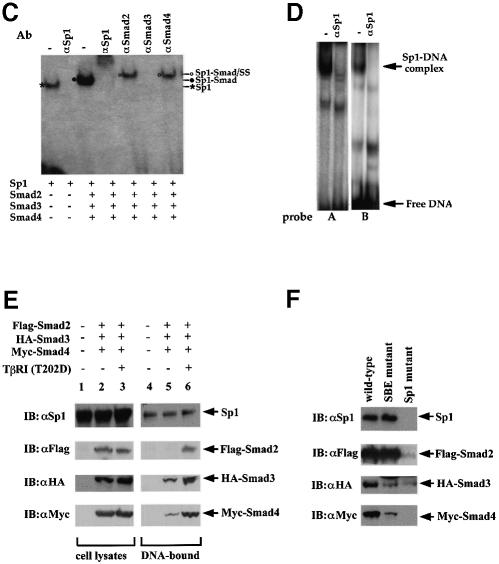
Fig. 5. Interaction of Smad2 or Smad4 with Sp1 at the Sp1a and Sp1b binding sites. Purified Sp1 (0.5 U) and GST–Smad fusion proteins (0.2 µg) were incubated with the 32P-labeled probe A, containing the Sp1a binding site (A), or probe B, containing the Sp1b binding site (B) of the p15Ink4B promoter. DNA-bound Sp1 and Sp1–Smad complexes are marked. (C) Gel shift analyses using oligonucleotide A and purified proteins were carried out as in (A). Antibodies, shown above the gel, were added to the gel shift reactions and incubated for 90 min, prior to gel analysis. Sp1–DNA, Sp1–Smad–DNA complex, and the supershifted (SS) complexes are marked. (D) Sp1-binding ability of p15 promoter elements. Nuclear extracts from HaCaT cells were incubated with probe A or B in gel shift reactions, with or without anti-Sp1 antibody. The Sp1–DNA complex, which is displaced in the presence of anti-Sp1 antibody, is indicated. (E) TGF-β-induced formation of Smad–DNA complex in cell lysates. Expression plasmids for Flag–Smad2, HA–Smad3 and Myc–Smad4 were transfected. Forty-eight hours after transfection, cells were lysed and incubated with streptavidin paramagnetic beads (Dynal) with immobilized biotinylated p15 promoter DNA oligonucleotide (nt –84 to –46). After extensive washing, DNA-bound proteins were detected by SDS–PAGE followed by western blotting using the indicated antibodies (lanes 4–6). Immunoblotting of the cell lysates in parallel demonstrates the expression level of endogenous Sp1 and transfected Smads. (F) The TGF-β-induced formation of Smad–DNA complex in cell lysates is largely dependent on intact Sp1 binding sites. Binding of Sp1 and Smad2, Smad3, Smad4 and Sp1 to the wild-type oligonucleotide, as was also done in (E), was compared with their binding to the corresponding mutant oligonucleotides, in which the SBEs and Sp1 binding sequences were mutated, as shown in Figure 3A.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.