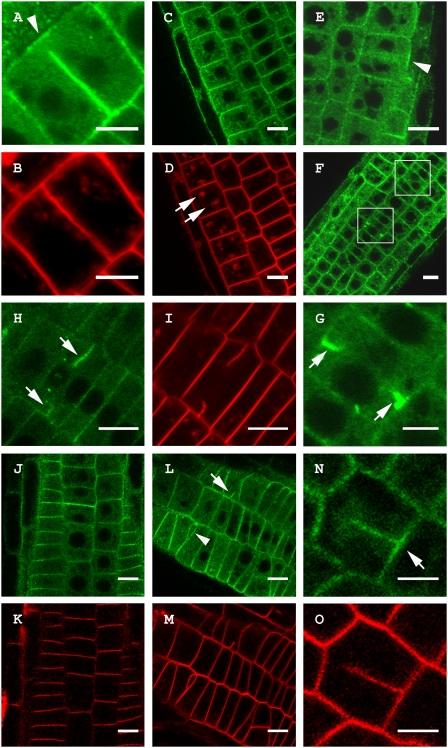
Fig. 4.
TPLATE-GFP accumulates at cell wall stubs in the Arabidopsis gnl1-1 mutant treated with BFA similar to Concanamycin A treatment but localization is not altered in the cpi1-1 background. (A) Untreated Arabidopsis root cell showing specific recruitment of TPLATE-GFP to the CDZ during cell plate anchoring independent of cell plate recruitment (arrowhead). (B) FM4-64 staining of the cell in A showing that the plate has contacted the PM and that the TPLATE-GFP label at the cell plate is absent from the outer rim of the plate. (C and D) Single section through a root expressing a genomic TPLATE-GFP treated with BFA (50 μM) and stained with FM4-64. TPLATE remains present at the PM and does not accumulate in BFA bodies (arrows in D). (E–G) Root epidermal cells of a gnl1-1 seedling expressing a genomic construct of TPLATE-GFP treated with 25 μM BFA. (E) Single plane confocal section of a dividing cell. TPLATE localizes to the plasma membrane and CDZ (arrowhead) but is absent between the reforming daughter nuclei. (F and G) Overview and close-up of gnl1-1 root epidermal cells showing TPLATE accumulation at cell wall stubs (white boxes, arrows). (H and I) TPLATE accumulation at cell wall stubs (arrows) in seedlings treated with 2 μM Concanamycin A. (J–O) Localization of a genomic TPLATE-GFP fusion in the cpi1-1 background stained with FM4-64. (J and K) TPLATE localization in the heterozygous cpi1-1 background. (L–O) TPLATE-GFP in the cpi1-1 mutant background. The cpi1-1 mutant shows altered division planes and cell wall stubs (arrowhead and arrow in L), yet TPLATE localization remains present at the PM (L) and the CDZ (arrow in N) in this background. (Scale bars: A, B, G, N, and O, 5 μm; C–F and H–M, 10 μm).