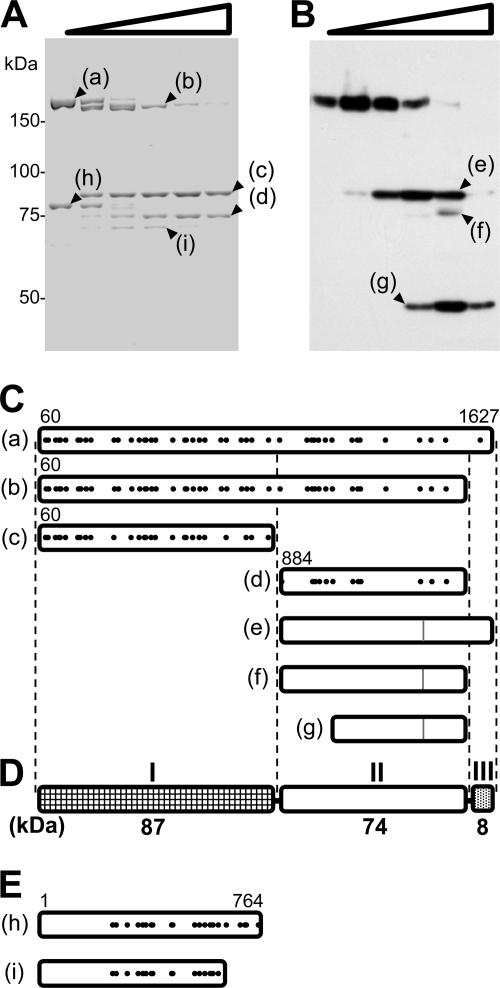
FIG. 4.
Partial digestion of the P1-P90 complex. (A) Protein profiles of the purified P1-P90 complex digested with various concentrations of trypsin. The protein fraction was treated with 0, 0.01, 0.03, 0.1, 0.3, and 1 mg/ml trypsin and subjected to SDS-8.75% PAGE. The resultant peptide fragments, marked a to i, were identified by PMF. (B) Immunoblot of M. pneumoniae cells treated with various concentrations of trypsin, using antibody against the C-terminal region of P1 adhesin for detection. The cells were treated with 0, 0.01, 0.03, 0.1, 0.3, and 1 mg/ml trypsin and subjected to SDS-PAGE and immunoblotting to detect the aa 1386 to 1394 sequence. The sizes of the resulting peptide fragments, marked e to g, were estimated to be 83, 77, and 48 kDa, respectively. The molecular masses are indicated on the left of panel A. (C) Mapping of products on the amino acid sequence of P1 adhesin. The 59 N-terminal residues predicted from the DNA sequence were processed in the mature form (46, 60). Protein bands a to d were identified by PMF. The positions of peptides detected in PMF are marked with solid dots. The N-terminal residues in bands a to d were determined by Edman analysis. The C terminus was estimated from the band size. The evident residue numbers of the ends are indicated. Peptide bands e to g were assigned from the band sizes, and the epitope positions are shown by gray lines. (D) Schematic of P1 adhesin, consisting of three domains, I, II, and III. The molecular mass is indicated below each domain. (E) Bands h and i mapped onto the amino acid sequence of P90, based on the results of PMF.